Genetic Diversity, Population Structure, and Andean Introgression in Brazilian Common Bean Cultivars after Half a Century of Genetic Breeding
- PMID: 33143347
- PMCID: PMC7694079
- DOI: 10.3390/genes11111298
Genetic Diversity, Population Structure, and Andean Introgression in Brazilian Common Bean Cultivars after Half a Century of Genetic Breeding
Abstract
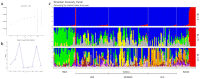
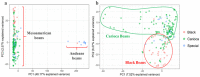
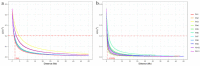
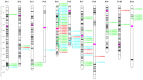
Brazil is the largest consumer and third highest producer of common beans (Phaseolus vulgaris L.) worldwide. Since the 1980s, the commercial Carioca variety has been the most consumed in Brazil, followed by Black and Special beans. The present study evaluates genetic diversity and population structure of 185 Brazilian common bean cultivars using 2827 high-quality single-nucleotide polymorphisms (SNPs). The Andean allelic introgression in the Mesoamerican accessions was investigated, and a Carioca panel was tested using an association mapping approach. The results distinguish the Mesoamerican from the Andean accessions, with a prevalence of Mesoamerican accessions (94.6%). When considering the commercial classes, low levels of genetic differentiation were seen, and the Carioca group showed the lowest genetic diversity. However, gain in gene diversity and allelic richness was seen for the modern Carioca cultivars. A set of 1060 'diagnostic SNPs' that show alternative alleles between the pure Mesoamerican and Andean accessions were identified, which allowed the identification of Andean allelic introgression events and shows that there are putative introgression segments in regions enriched with resistance genes. Finally, genome-wide association studies revealed SNPs significantly associated with flowering time, pod maturation, and growth habit, showing that the Carioca Association Panel represents a powerful tool for crop improvements.
Keywords: Carioca variety; Phaseolus vulgaris L.; common bean; genome-wide association study; genomic introgression; variability gain.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Broughton W.J., Hernández G., Blair M., Beebe S., Gepts P., Vanderleyden J. Beans (Phaseolus spp.)—Model food legumes. Plant Soil. 2003;252:55–128.
-
- Bellucci E., Bitocchi E., Rau D., Rodriguez M., Biagetti E., Giardini A., Attene G., Nanni L., Papa R. Genomics of origin, domestication and evolution of Phaseolus vulgaris. In: Tuberosa R., Graner A., Frison E., editors. Genomics of Plant Genetic Resources. Springer Netherlands; Dordrecht, The Netherlands: 2014.
-
- Pires C.V., Oliveira M.G.A., Cruz G.A.D.R., Mendes F.Q., De Rezende S.T., Moreira M.A. Composição físico-química de diferentes cultivares de feijão (Phaseolus vulgaris L.) Alimentos e Nutricao Araraquara. 2005;16:157–162.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

