Identification of natural inhibitors against Mpro of SARS-CoV-2 by molecular docking, molecular dynamics simulation, and MM/PBSA methods
- PMID: 33143552
- PMCID: PMC7651194
- DOI: 10.1080/07391102.2020.1842806
Identification of natural inhibitors against Mpro of SARS-CoV-2 by molecular docking, molecular dynamics simulation, and MM/PBSA methods
Abstract
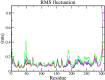
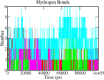
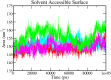
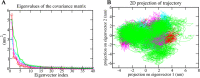
The recent outbreak of SARS-CoV-2 disease, also known as COVID-19, has emerged as a pandemic. The unavailability of specific therapeutic drugs and vaccines urgently demands sincere efforts for drug discovery against COVID-19. The main protease (Mpro) of SARS-CoV-2 is a critical drug target as it plays an essential role in virus replication. Therefore for the identification of potential inhibitors of SARS-CoV-2 Mpro, we applied a structure-based virtual screening approach followed by molecular dynamics (MD) study. A library of 686 phytochemicals was subjected to virtual screening which resulted in 28 phytochemicals based on binding energy. These phytochemicals were further subjected to drug-likeness and toxicity analysis, which resulted in seven drug-like hits. Out of seven, five phytochemicals viz., Mpro-Dehydrtectol (-10.3 kcal/mol), Epsilon-viniferin (-8.6 kcal/mol), Peimisine (-8.6 kcal/mol), Gmelanone (-8.4 kcal/mol), and Isocolumbin (-8.4 kcal/mol) were non-toxic. Consequently, these phytochemicals are subjected to MD, post MD analysis, and MM/PBSA calculations. The results of 100 ns MD simulation, RMSF, SASA, Rg, and MM/PBSA show that Epsilon-viniferin (-29.240 kJ/mol), Mpro-Peimisine (-43.031 kJ/mol) and Gmelanone (-13.093 kJ/mol) form a stable complex with Mpro and could be used as potential inhibitors of SARS-CoV-2 Mpro. However, further investigation of these inhibitors against Mpro receptor of COVID-19 is needed to validate their candidacy for clinical trials. Communicated by Ramaswamy H. Sarma.
Keywords: COVID-19; Coronavirus; Mpro; molecular docking; molecular dynamic simulation.
Conflict of interest statement
The authors declare that there is no competing interest in this work.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
