Treatment- and population-specific genetic risk factors for anti-drug antibodies against interferon-beta: a GWAS
- PMID: 33143745
- PMCID: PMC7641861
- DOI: 10.1186/s12916-020-01769-6
Treatment- and population-specific genetic risk factors for anti-drug antibodies against interferon-beta: a GWAS
Abstract
Background: Upon treatment with biopharmaceuticals, the immune system may produce anti-drug antibodies (ADA) that inhibit the therapy. Up to 40% of multiple sclerosis patients treated with interferon β (IFNβ) develop ADA, for which a genetic predisposition exists. Here, we present a genome-wide association study on ADA and predict the occurrence of antibodies in multiple sclerosis patients treated with different interferon β preparations.
Methods: We analyzed a large sample of 2757 genotyped and imputed patients from two cohorts (Sweden and Germany), split between a discovery and a replication dataset. Binding ADA (bADA) levels were measured by capture-ELISA, neutralizing ADA (nADA) titers using a bioassay. Genome-wide association analyses were conducted stratified by cohort and treatment preparation, followed by fixed-effects meta-analysis.
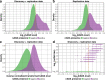
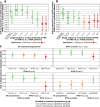
Results: Binding ADA levels and nADA titers were correlated and showed a significant heritability (47% and 50%, respectively). The risk factors differed strongly by treatment preparation: The top-associated and replicated variants for nADA presence were the HLA-associated variants rs77278603 in IFNβ-1a s.c.- (odds ratio (OR) = 3.55 (95% confidence interval = 2.81-4.48), p = 2.1 × 10-26) and rs28366299 in IFNβ-1b s.c.-treated patients (OR = 3.56 (2.69-4.72), p = 6.6 × 10-19). The rs77278603-correlated HLA haplotype DR15-DQ6 conferred risk specifically for IFNβ-1a s.c. (OR = 2.88 (2.29-3.61), p = 7.4 × 10-20) while DR3-DQ2 was protective (OR = 0.37 (0.27-0.52), p = 3.7 × 10-09). The haplotype DR4-DQ3 was the major risk haplotype for IFNβ-1b s.c. (OR = 7.35 (4.33-12.47), p = 1.5 × 10-13). These haplotypes exhibit large population-specific frequency differences. The best prediction models were achieved for ADA in IFNβ-1a s.c.-treated patients. Here, the prediction in the Swedish cohort showed AUC = 0.91 (0.85-0.95), sensitivity = 0.78, and specificity = 0.90; patients with the top 30% of genetic risk had, compared to patients in the bottom 30%, an OR = 73.9 (11.8-463.6, p = 4.4 × 10-6) of developing nADA. In the German cohort, the AUC of the same model was 0.83 (0.71-0.92), sensitivity = 0.80, specificity = 0.76, with an OR = 13.8 (3.0-63.3, p = 7.5 × 10-4).
Conclusions: We identified several HLA-associated genetic risk factors for ADA against interferon β, which were specific for treatment preparations and population backgrounds. Genetic prediction models could robustly identify patients at risk for developing ADA and might be used for personalized therapy recommendations and stratified ADA screening in clinical practice. These analyses serve as a roadmap for genetic characterizations of ADA against other biopharmaceutical compounds.
Keywords: Anti-drug antibodies; Genetics; Genome-wide association study; Human leukocyte antigen (HLA) system; Interferon beta; Multiple sclerosis; Prediction.
Conflict of interest statement
TFMA, JL, DM, MR, VG, LA, CG, PEHJ, IK, and MP have no competing interests to declare.
CH is an employee of Sanofi Genzyme.
MA has received speaker honoraria and/or travel grants from Biogen, Novartis, Merck, and Sanofi Genzyme.
HH has participated in meetings sponsored by and received speaker honoraria or travel funding from Bayer, Biogen, Merck, Novartis, Sanofi-Genzyme, Siemens, and Teva, and received honoraria for acting as consultant for Biogen and Teva.
BK received a research grant and travel compensations from Novartis outside the submitted work.
FS has served on scientific advisory boards, been on the steering committees of clinical trials, served as a consultant, received support for congress participation, received speaker honoraria, or received research support for his laboratory from Biogen, Merck, Novartis, Roche, Sanofi Genzyme, and Teva.
TO has received unrestricted MS research grants, and honoraria for advisory boards/lectures from Biogen, Novartis, Sanofi, Merck, and Roche.
SeS is a former employee and has stocks and/or stock options in Novartis.
FD has participated in meetings sponsored by or received honoraria for acting as an advisor/speaker for Almirall, Alexion, Biogen, Celgene, Genzyme-Sanofi, Merck, Novartis Pharma, Roche, and TEVA ratiopharm. His institution has received research grants from Biogen and Genzyme Sanofi. He is section editor of the MSARD Journal (Multiple Sclerosis and Related Disorders).
AFH has received unrestricted research grants from Merck-Serono and BiogenIdec, served as consultant for Johnson & Johnson, and received honoraria for lectures by BiogenIdec and Sanofi-Aventis.
During the last 2 years, BH has served on scientific advisory boards for Novartis; he has served as DMSC member for AllergyCare, Polpharma, and TG therapeutics; he or his institution have received speaker honoraria from Desitin; his institution received research grants from Regeneron for MS research; he holds part of a patent for the detection of antibodies against KIR4.1 in a subpopulation of patients with MS. None of these conflicts are relevant to the topic of the study.
BMM and BH hold parts of a patent for genetic determinants of neutralizing antibodies to interferon.
Figures

References
-
- Bertolotto A, Deisenhammer F, Gallo P, Sørensen P. Immunogenicity of interferon beta: differences among products. J Neurol. 2004;251(Suppl 2):ii15–ii24. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 115303/Innovative Medicines Initiative Joint Undertaking/International
- 01ZZ1804A/Bundesministerium für Bildung und Forschung/International
- 01ZX1614J/Bundesministerium für Bildung und Forschung/International
- 733161/H2020 European Research Council/International
- GA 2913/1-1/Deutsche Forschungsgemeinschaft/International
LinkOut - more resources
Full Text Sources
Research Materials

