Changes in SARS-CoV-2 Spike versus Nucleoprotein Antibody Responses Impact the Estimates of Infections in Population-Based Seroprevalence Studies
- PMID: 33144321
- PMCID: PMC7925109
- DOI: 10.1128/JVI.01828-20
Changes in SARS-CoV-2 Spike versus Nucleoprotein Antibody Responses Impact the Estimates of Infections in Population-Based Seroprevalence Studies
Abstract
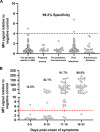
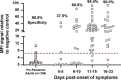
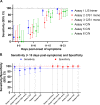
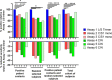
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-specific antibody responses to the spike (S) protein monomer, S protein native trimeric form, or the nucleocapsid (N) proteins were evaluated in cohorts of individuals with acute infection (n = 93) and in individuals enrolled in a postinfection seroprevalence population study (n = 578) in Switzerland. Commercial assays specific for the S1 monomer, for the N protein, or within a newly developed Luminex assay using the S protein trimer were found to be equally sensitive in antibody detection in the acute-infection-phase samples. Interestingly, compared to anti-S antibody responses, those against the N protein appear to wane in the postinfection cohort. Seroprevalence in a "positive patient contacts" group (n = 177) was underestimated by N protein assays by 10.9 to 32.2%, while the "randomly selected" general population group (n = 311) was reduced by up to 45% relative to the S protein assays. The overall reduction in seroprevalence targeting only anti-N antibodies for the total cohort ranged from 9.4 to 31%. Of note, the use of the S protein in its native trimer form was significantly more sensitive compared to monomeric S proteins. These results indicate that the assessment of anti-S IgG antibody responses against the native trimeric S protein should be implemented to estimate SARS-CoV-2 infections in population-based seroprevalence studies.IMPORTANCE In the present study, we have determined SARS-CoV-2-specific antibody responses in sera of acute and postinfection phase subjects. Our results indicate that antibody responses against viral S and N proteins were equally sensitive in the acute phase of infection, but that responses against N appear to wane in the postinfection phase where those against the S protein persist over time. The most sensitive serological assay in both acute and postinfection phases used the native S protein trimer as the binding antigen, which has significantly greater conformational epitopes for antibody binding compared to the S1 monomer protein used in other assays. We believe these results are extremely important in order to generate correct estimates of SARS-CoV-2 infections in the general population. Furthermore, the assessment of antibody responses against the trimeric S protein will be critical to evaluate the durability of the antibody response and for the characterization of a vaccine-induced antibody response.
Keywords: S protein trimer; SARS-CoV-2; serology.
Copyright © 2021 Fenwick et al.
Figures

References
-
- World Health Organization. 2020. Coronavirus disease (COVID-2019) situation reports-175. World Health Organization, Geneva, Switzerland.
-
- Okba NMA, Muller MA, Li W, Wang C, GeurtsvanKessel CH, Corman VM, Lamers MM, Sikkema RS, de Bruin E, Chandler FD, Yazdanpanah Y, Le Hingrat Q, Descamps D, Houhou-Fidouh N, Reusken C, Bosch BJ, Drosten C, Koopmans MPG, Haagmans BL. 2020. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg Infect Dis 26:1478–1488. doi: 10.3201/eid2607.200841. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

