Adaptation of influenza viruses to human airway receptors
- PMID: 33144323
- PMCID: PMC7948470
- DOI: 10.1074/jbc.REV120.013309
Adaptation of influenza viruses to human airway receptors
Abstract
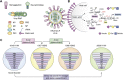
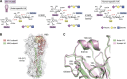
Through annual epidemics and global pandemics, influenza A viruses (IAVs) remain a significant threat to human health as the leading cause of severe respiratory disease. Within the last century, four global pandemics have resulted from the introduction of novel IAVs into humans, with components of each originating from avian viruses. IAVs infect many avian species wherein they maintain a diverse natural reservoir, posing a risk to humans through the occasional emergence of novel strains with enhanced zoonotic potential. One natural barrier for transmission of avian IAVs into humans is the specificity of the receptor-binding protein, hemagglutinin (HA), which recognizes sialic-acid-containing glycans on host cells. HAs from human IAVs exhibit "human-type" receptor specificity, binding exclusively to glycans on cells lining the human airway where terminal sialic acids are attached in the α2-6 configuration (NeuAcα2-6Gal). In contrast, HAs from avian viruses exhibit specificity for "avian-type" α2-3-linked (NeuAcα2-3Gal) receptors and thus require adaptive mutations to bind human-type receptors. Since all human IAV pandemics can be traced to avian origins, there remains ever-present concern over emerging IAVs with human-adaptive potential that might lead to the next pandemic. This concern has been brought into focus through emergence of SARS-CoV-2, aligning both scientific and public attention to the threat of novel respiratory viruses from animal sources. In this review, we summarize receptor-binding adaptations underlying the emergence of all prior IAV pandemics in humans, maintenance and evolution of human-type receptor specificity in subsequent seasonal IAVs, and potential for future human-type receptor adaptation in novel avian HAs.
Keywords: cell surface receptor; glycoprotein; hemagglutinin; influenza virus; neuraminidase; pandemic; sialic acid.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures


References
-
- Shinya K., Ebina M., Yamada S., Ono M., Kasai N., Kawaoka Y. Avian flu: influenza virus receptors in the human airway. Nature. 2006;440:435–436. - PubMed
-
- Rogers G.N., Paulson J.C. Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology. 1983;127:361–373. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

