Nearest-neighbor NMR spectroscopy: categorizing spectral peaks by their adjacent nuclei
- PMID: 33144564
- PMCID: PMC7642304
- DOI: 10.1038/s41467-020-19325-4
Nearest-neighbor NMR spectroscopy: categorizing spectral peaks by their adjacent nuclei
Abstract
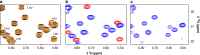
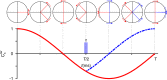
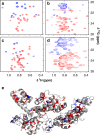
Methyl-NMR enables atomic-resolution studies of structure and dynamics of large proteins in solution. However, resonance assignment remains challenging. The problem is to combine existing structural informational with sparse distance restraints and search for the most compatible assignment among the permutations. Prior classification of peaks as either from isoleucine, leucine, or valine reduces the search space by many orders of magnitude. However, this is hindered by overlapped leucine and valine frequencies. In contrast, the nearest-neighbor nuclei, coupled to the methyl carbons, resonate in distinct frequency bands. Here, we develop a framework to imprint additional information about passively coupled resonances onto the observed peaks. This depends on simultaneously orchestrating closely spaced bands of resonances along different magnetization trajectories, using principles from control theory. For methyl-NMR, the method is implemented as a modification to the standard fingerprint spectrum (the 2D-HMQC). The amino acid type is immediately apparent in the fingerprint spectrum. There is no additional relaxation loss or an increase in experimental time. The method is validated on biologically relevant proteins. The idea of generating new spectral information using passive, adjacent resonances is applicable to other contexts in NMR spectroscopy.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

