Rampant loss of social traits during domestication of a Bacillus subtilis natural isolate
- PMID: 33144634
- PMCID: PMC7642357
- DOI: 10.1038/s41598-020-76017-1
Rampant loss of social traits during domestication of a Bacillus subtilis natural isolate
Abstract
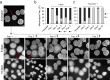
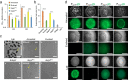
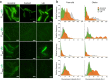
Most model bacteria have been domesticated in laboratory conditions. Yet, the tempo with which a natural isolate diverges from its ancestral phenotype under domestication to a novel laboratory environment is poorly understood. Such knowledge, however is essential to understanding the rate of evolution, the time scale over which a natural isolate can be propagated without loss of its natural adaptive traits, and the reliability of experimental results across labs. Using experimental evolution, phenotypic assays, and whole-genome sequencing, we show that within a week of propagation in a common laboratory environment, a natural isolate of Bacillus subtilis acquires mutations that cause changes in a multitude of traits. A single adaptive mutational step in the gene coding for the transcriptional regulator DegU impairs a DegU-dependent positive autoregulatory loop and leads to loss of robust biofilm architecture, impaired swarming motility, reduced secretion of exoproteases, and to changes in the dynamics of sporulation across environments. Importantly, domestication also resulted in improved survival when the bacteria face pressure from cells of the innate immune system. These results show that degU is a target for mutations during domestication and underscores the importance of performing careful and extremely short-term propagations of natural isolates to conserve the traits encoded in their original genomes.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

