Identification of Differential Intestinal Mucosa Transcriptomic Biomarkers for Ulcerative Colitis by Bioinformatics Analysis
- PMID: 33144895
- PMCID: PMC7596466
- DOI: 10.1155/2020/8876565
Identification of Differential Intestinal Mucosa Transcriptomic Biomarkers for Ulcerative Colitis by Bioinformatics Analysis
Abstract
Background: Ulcerative colitis (UC) is a complicated disease caused by the interaction between genetic and environmental factors that affect mucosal homeostasis and triggers inappropriate immune response. The purpose of the study was to identify significant biomarkers with potential therapeutic targets and the underlying mechanisms.
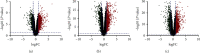
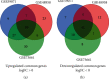
Methods: The gene expression profiles of GSE48958, GSE73661, and GSE59071 are from the GEO database. Differentially expressed genes (DEGs) were screened by the GEO2R tool. Next, the Database for Annotation, Visualization and Integrated Discovery (DAVID) was applied to analyze gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway. Then, protein-protein interaction (PPI) was visualized by Cytoscape with Search Tool for the Retrieval of Interacting Genes (STRING).
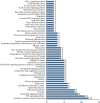
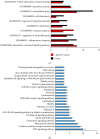
Results: There were a total of 128 common DEGs genes, including 86 upregulated genes enriched in extracellular space, regulation of inflammatory response, chemokine-mediated signaling pathway, response to lipopolysaccharide, and cell proliferation, while 42 downregulated genes enriched in the integral component of the membrane, the integral component of the plasma membrane, apical plasma membrane, symporter activity, and chloride channel activity. The KEGG pathway analysis results demonstrated that DEGs were particularly enriched in cytokine-cytokine receptor interaction, TNF signaling pathway, chemokine signaling pathway, pertussis, and rheumatoid arthritis. 18 central modules of the PPI networks were selected with Cytotype MCODE. Furthermore, 18 genes were found to significantly enrich in the extracellular space, inflammatory response, chemokine-mediated signaling pathway, TNF signaling pathway, regulation of cell proliferation, and immune response via reanalysis of DAVID.
Conclusion: The study identified DEGs, key target genes, functional pathways, and pathway analysis of UC, which may provide potential molecular targets and diagnostic biomarkers for UC.
Copyright © 2020 Fang Cheng et al.
Conflict of interest statement
All authors report no conflicts of interest.
Figures


Similar articles
-
Identification of differentially expressed genes in ulcerative colitis and verification in a colitis mouse model by bioinformatics analyses.World J Gastroenterol. 2020 Oct 21;26(39):5983-5996. doi: 10.3748/wjg.v26.i39.5983. World J Gastroenterol. 2020. PMID: 33132649 Free PMC article.
-
Identification of significant genes as prognostic markers and potential tumor suppressors in lung adenocarcinoma via bioinformatical analysis.BMC Cancer. 2021 May 26;21(1):616. doi: 10.1186/s12885-021-08308-3. BMC Cancer. 2021. PMID: 34039311 Free PMC article.
-
Bioinformatics analysis of fibroblasts exposed to TGF‑β at the early proliferation phase of wound repair.Mol Med Rep. 2017 Dec;16(6):8146-8154. doi: 10.3892/mmr.2017.7619. Epub 2017 Sep 26. Mol Med Rep. 2017. PMID: 28983581 Free PMC article.
-
Effects of dietary curcumin on gene expression: An analysis of transcriptomic data in mice.Pathol Res Pract. 2024 Nov;263:155653. doi: 10.1016/j.prp.2024.155653. Epub 2024 Oct 11. Pathol Res Pract. 2024. PMID: 39426142 Review.
-
Identification of Potential Drug Therapy for Dermatofibrosarcoma Protuberans with Bioinformatics and Deep Learning Technology.Curr Comput Aided Drug Des. 2022;18(5):393-405. doi: 10.2174/1573409918666220816112206. Curr Comput Aided Drug Des. 2022. PMID: 35975851 Review.
Cited by
-
Establishment of potential lncRNA-related hub genes involved competitive endogenous RNA in lung adenocarcinoma.BMC Cancer. 2024 Nov 9;24(1):1371. doi: 10.1186/s12885-024-13144-2. BMC Cancer. 2024. PMID: 39522011 Free PMC article.
-
Identification and immune landscape of sarcopenia-related molecular clusters in inflammatory bowel disease by machine learning and integrated bioinformatics.Sci Rep. 2024 Jul 30;14(1):17603. doi: 10.1038/s41598-024-68198-w. Sci Rep. 2024. PMID: 39079987 Free PMC article.
-
Identification of potential diagnostic markers and molecular mechanisms of asthma and ulcerative colitis based on bioinformatics and machine learning.Front Mol Biosci. 2025 May 15;12:1554304. doi: 10.3389/fmolb.2025.1554304. eCollection 2025. Front Mol Biosci. 2025. PMID: 40443529 Free PMC article.
-
Integrated Analysis of Multiple Microarray Studies to Identify Novel Gene Signatures in Ulcerative Colitis.Front Genet. 2021 Jul 9;12:697514. doi: 10.3389/fgene.2021.697514. eCollection 2021. Front Genet. 2021. PMID: 34306038 Free PMC article.
-
TDAG51 deficiency attenuates dextran sulfate sodium-induced colitis in mice.Sci Rep. 2022 Nov 30;12(1):20619. doi: 10.1038/s41598-022-24873-4. Sci Rep. 2022. PMID: 36450854 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

