The insertion of a mitochondrial selfish element into the nuclear genome and its consequences
- PMID: 33144953
- PMCID: PMC7593156
- DOI: 10.1002/ece3.6749
The insertion of a mitochondrial selfish element into the nuclear genome and its consequences
Abstract
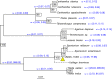
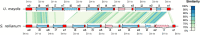
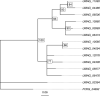
Homing endonucleases (HE) are enzymes capable of cutting DNA at highly specific target sequences, the repair of the generated double-strand break resulting in the insertion of the HE-encoding gene ("homing" mechanism). HEs are present in all three domains of life and viruses; in eukaryotes, they are mostly found in the genomes of mitochondria and chloroplasts, as well as nuclear ribosomal RNAs. We here report the case of a HE that accidentally integrated into a telomeric region of the nuclear genome of the fungal maize pathogen Ustilago maydis. We show that the gene has a mitochondrial origin, but its original copy is absent from the U. maydis mitochondrial genome, suggesting a subsequent loss or a horizontal transfer from a different species. The telomeric HE underwent mutations in its active site and lost its original start codon. A potential other start codon was retained downstream, but we did not detect any significant transcription of the newly created open reading frame, suggesting that the inserted gene is not functional. Besides, the insertion site is located in a putative RecQ helicase gene, truncating the C-terminal domain of the protein. The truncated helicase is expressed during infection of the host, together with other homologous telomeric helicases. This unusual mutational event altered two genes: The integrated HE gene subsequently lost its homing activity, while its insertion created a truncated version of an existing gene, possibly altering its function. As the insertion is absent in other field isolates, suggesting that it is recent, the U. maydis 521 reference strain offers a snapshot of this singular mutational event.
Keywords: gene birth; gene transfer; homing endonuclease; intron; mitochondrion.
© 2020 The Authors. Ecology and Evolution published by John Wiley & Sons Ltd.
Conflict of interest statement
None declared.
Figures




Similar articles
-
The mitochondrial LSU rRNA group II intron of Ustilago maydis encodes an active homing endonuclease likely involved in intron mobility.PLoS One. 2012;7(11):e49551. doi: 10.1371/journal.pone.0049551. Epub 2012 Nov 14. PLoS One. 2012. PMID: 23166709 Free PMC article.
-
Organization of chromosome ends in Ustilago maydis. RecQ-like helicase motifs at telomeric regions.Genetics. 1998 Mar;148(3):1043-54. doi: 10.1093/genetics/148.3.1043. Genetics. 1998. PMID: 9539423 Free PMC article.
-
I-OmiI and I-OmiII: two intron-encoded homing endonucleases within the Ophiostoma minus rns gene.Fungal Biol. 2014 Aug;118(8):721-31. doi: 10.1016/j.funbio.2014.05.002. Epub 2014 May 23. Fungal Biol. 2014. PMID: 25110134
-
Structural and functional characteristics of homing endonucleases.Crit Rev Biochem Mol Biol. 2003;38(3):199-248. doi: 10.1080/713609235. Crit Rev Biochem Mol Biol. 2003. PMID: 12870715 Review.
-
Ancient endosymbiont-mediated transmission of a selfish gene provides a model for overcoming barriers to gene transfer into animal mitochondrial genomes.Bioessays. 2023 Feb;45(2):e2200190. doi: 10.1002/bies.202200190. Epub 2022 Nov 22. Bioessays. 2023. PMID: 36412071 Review.
Cited by
-
First characterization of the complete mitochondrial genome of fungal plant-pathogen Monilinia laxa which represents the mobile intron rich structure.Sci Rep. 2020 Aug 12;10(1):13644. doi: 10.1038/s41598-020-70611-z. Sci Rep. 2020. PMID: 32788650 Free PMC article.
-
First Characterization of a Hafnia Phage Reveals Extraordinarily Large Burst Size and Unusual Plaque Polymorphism.Front Microbiol. 2022 Feb 8;12:754331. doi: 10.3389/fmicb.2021.754331. eCollection 2021. Front Microbiol. 2022. PMID: 35211099 Free PMC article.
-
Global Gene Expression of Post-Senescent Telomerase-Negative ter1Δ Strain of Ustilago maydis.J Fungi (Basel). 2023 Aug 31;9(9):896. doi: 10.3390/jof9090896. J Fungi (Basel). 2023. PMID: 37755003 Free PMC article.
-
Exploring Mitochondrial Heterogeneity and Evolutionary Dynamics in Thelephora ganbajun through Population Genomics.Int J Mol Sci. 2024 Aug 19;25(16):9013. doi: 10.3390/ijms25169013. Int J Mol Sci. 2024. PMID: 39201699 Free PMC article.
-
Comparative mitogenomic analysis of Sporisorium reilianum f. sp. zeae suggests recombination events during its evolutionary history.Front Physiol. 2024 Sep 6;15:1264359. doi: 10.3389/fphys.2024.1264359. eCollection 2024. Front Physiol. 2024. PMID: 39308980 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

