Investigation of hub genes involved in diabetic nephropathy using biological informatics methods
- PMID: 33145306
- PMCID: PMC7575993
- DOI: 10.21037/atm-20-5647
Investigation of hub genes involved in diabetic nephropathy using biological informatics methods
Abstract
Background: The aim of this study was to find genes with significantly aberrant expression in diabetic nephropathy (DN) and determine their underlying mechanisms.
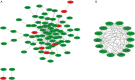
Methods: GSE30528 and GSE1009 were obtained by querying the Gene Expression Omnibus (GEO) database. The difference in target gene expression between normal renal tissues and kidney tissues in patients with DN was screened by using the GEO2R tool. Using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) database, differentially expressed genes (DEGs) were analysed by Gene Ontology (GO) annotation and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment. Then, the protein-protein interactions (PPIs) of DEGs were analyzed by Cytoscape with the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database, and the hub genes in this PPI network were recognized by centrality analysis.
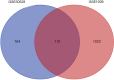
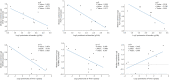
Results: There were 110 genes with significant expression differences between normal and DN tissues. The differences in gene expression involved many functions and expression pathways, such as the formation of the extracellular matrix and the construction of the extracellular domain. The correlation analysis and subgroup analysis of 14 hub genes and the clinical characteristics of DN showed that CTGF, ALB, PDPN, FLT1, IGF1, WT1, GJA1, IGFBP2, FGF9, BMP2, FGF1, BMP7, VEGFA, and TGFBR3 may be involved in the progression of DN.
Conclusions: We confirmed the differentially expressed hub genes and other genes which may be the novel biomarker and target candidates in DN.
Keywords: Diabetic nephropathy (DN); bioinformatics analysis; differentially expressed gene (DEG); hub genes; microarray analysis.
2020 Annals of Translational Medicine. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-5647). The authors have no conflicts of interest to declare.
Figures


References
LinkOut - more resources
Full Text Sources
Miscellaneous
