Designer Fluorescent Adenines Enable Real-Time Monitoring of MUTYH Activity
- PMID: 33145410
- PMCID: PMC7596860
- DOI: 10.1021/acscentsci.0c00369
Designer Fluorescent Adenines Enable Real-Time Monitoring of MUTYH Activity
Abstract
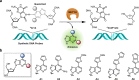
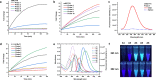
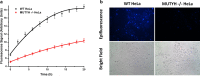
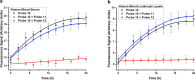
The human DNA base excision repair enzyme MUTYH (MutY homolog DNA glycosylase) excises undamaged adenine that has been misincorporated opposite the oxidatively damaged 8-oxoG, preventing transversion mutations and serving as an important defense against the deleterious effects of this damage. Mutations in the MUTYH gene predispose patients to MUTYH-associated polyposis and colorectal cancer, and MUTYH expression has been documented as a biomarker for pancreatic cancer. Measuring MUTYH activity is therefore critical for evaluating and diagnosing disease states as well as for testing this enzyme as a potential therapeutic target. However, current methods for measuring MUTYH activity rely on indirect electrophoresis and radioactivity assays, which are difficult to implement in biological and clinical settings. Herein, we synthesize and identify novel fluorescent adenine derivatives that can act as direct substrates for excision by MUTYH as well as bacterial MutY. When incorporated into synthetic DNAs, the resulting fluorescently modified adenine-release turn-on (FMART) probes report on enzymatic base excision activity in real time, both in vitro and in mammalian cells and human blood. We also employ the probes to identify several promising small-molecule modulators of MUTYH by employing FMART probes for in vitro screening.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
-
- Al-Tassan N.; Chmiel N. H.; Maynard J.; Fleming N.; Livingston A. L.; Williams G. T.; Hodges A. K.; Davies D. R.; David S. S.; Sampson J. R.; Cheadle J. P. Inherited variants of MYH associated with somatic G:C-->T:A mutations in colorectal tumors. Nat. Genet. 2002, 30 (2), 227–232. 10.1038/ng828. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

