Synthetic Elaboration of Native DNA by RASS (SENDR)
- PMID: 33145415
- PMCID: PMC7596865
- DOI: 10.1021/acscentsci.0c00680
Synthetic Elaboration of Native DNA by RASS (SENDR)
Erratum in
-
Correction to "Synthetic Elaboration of Native DNA by RASS (SENDR)".ACS Cent Sci. 2020 Nov 25;6(11):2117. doi: 10.1021/acscentsci.0c01389. Epub 2020 Oct 23. ACS Cent Sci. 2020. PMID: 33274288 Free PMC article.
Abstract
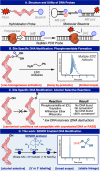
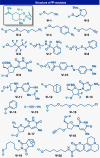
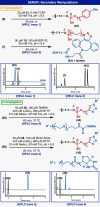
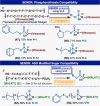
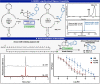
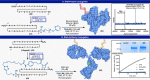
Controlled site-specific bioconjugation through chemical methods to native DNA remains an unanswered challenge. Herein, we report a simple solution to achieve this conjugation through the tactical combination of two recently developed technologies: one for the manipulation of DNA in organic media and another for the chemoselective labeling of alcohols. Reversible adsorption of solid support (RASS) is employed to immobilize DNA and facilitate its transfer into dry acetonitrile. Subsequent reaction with P(V)-based Ψ reagents takes place in high yield with exquisite selectivity for the exposed 3' or 5' alcohols on DNA. This two-stage process, dubbed SENDR for Synthetic Elaboration of Native DNA by RASS, can be applied to a multitude of DNA conformations and sequences with a variety of functionalized Ψ reagents to generate useful constructs.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

