A Photodeactivatable Antagonist for Controlling CREB-Dependent Gene Expression
- PMID: 33145417
- PMCID: PMC7596873
- DOI: 10.1021/acscentsci.0c00736
A Photodeactivatable Antagonist for Controlling CREB-Dependent Gene Expression
Abstract
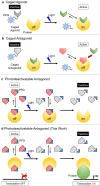
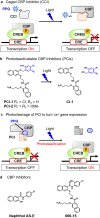
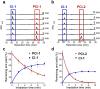
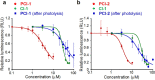
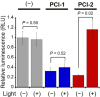
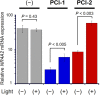
A novel photodeactivation strategy for controlling gene expression has been developed based on light-induced activation of cAMP response element binding protein (CREB). Light-induced cleavage of the photoresponsive protecting group of an antagonist of CREB binding protein (CBP) results in photocleaved products with weak binding affinity for CBP. This photodissociation reaction enables protein-protein interactions between CBP and CREB that trigger the formation of a multiprotein transcription complex to turn gene expression "on". This enables irradiation of antagonist-treated HEK293T cells to be used to trigger temporal recovery of CREB-dependent transcriptional activity and endogenous gene expression under photolytic control.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
Similar articles
-
Distinct effects of cAMP and mitogenic signals on CREB-binding protein recruitment impart specificity to target gene activation via CREB.Proc Natl Acad Sci U S A. 2001 Sep 11;98(19):10936-41. doi: 10.1073/pnas.191152098. Epub 2001 Sep 4. Proc Natl Acad Sci U S A. 2001. PMID: 11535812 Free PMC article.
-
The expanding family of CREB/CREM transcription factors that are involved with spermatogenesis.Mol Cell Endocrinol. 2002 Feb 22;187(1-2):115-24. doi: 10.1016/s0303-7207(01)00696-7. Mol Cell Endocrinol. 2002. PMID: 11988318 Review.
-
Role of the transcriptional coactivator CBP/p300 in linking basic helix-loop-helix and CREB responses for follicle-stimulating hormone-mediated activation of the transferrin promoter in Sertoli cells.Biol Reprod. 2001 Aug;65(2):568-74. doi: 10.1095/biolreprod65.2.568. Biol Reprod. 2001. PMID: 11466227
-
Cooperative interactions between CBP and TORC2 confer selectivity to CREB target gene expression.EMBO J. 2007 Jun 20;26(12):2880-9. doi: 10.1038/sj.emboj.7601715. Epub 2007 May 3. EMBO J. 2007. PMID: 17476304 Free PMC article.
-
Regulation of somatostatin gene transcription by cyclic adenosine monophosphate.Metabolism. 1996 Aug;45(8 Suppl 1):4-7. doi: 10.1016/s0026-0495(96)90068-2. Metabolism. 1996. PMID: 8769368 Review.
Cited by
-
Deactivatable Bisubstrate Inhibitors of Protein Kinases.Molecules. 2022 Oct 8;27(19):6689. doi: 10.3390/molecules27196689. Molecules. 2022. PMID: 36235226 Free PMC article.
-
Multi-faceted regulation of CREB family transcription factors.Front Mol Neurosci. 2024 Aug 6;17:1408949. doi: 10.3389/fnmol.2024.1408949. eCollection 2024. Front Mol Neurosci. 2024. PMID: 39165717 Free PMC article. Review.
References
-
- Yip R. W.; Wen Y. X.; Gravel D.; Giasson R.; Sharma D. K Photochemistry of the ortho-Nitrobenzyl System in Solution - Identification of the Biradical Intermediate in the Intramolecular Rearrangement. J. Phys. Chem. 1991, 95, 6078–6081. 10.1021/j100169a009. - DOI
LinkOut - more resources
Full Text Sources

