ACE2 and ACE: structure-based insights into mechanism, regulation and receptor recognition by SARS-CoV
- PMID: 33146371
- PMCID: PMC7642307
- DOI: 10.1042/CS20200899
ACE2 and ACE: structure-based insights into mechanism, regulation and receptor recognition by SARS-CoV
Abstract
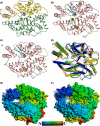
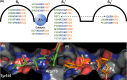
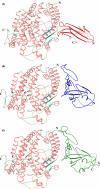
Angiotensin converting enzyme (ACE) is well-known for its role in blood pressure regulation via the renin-angiotensin aldosterone system (RAAS) but also functions in fertility, immunity, haematopoiesis and diseases such as obesity, fibrosis and Alzheimer's dementia. Like ACE, the human homologue ACE2 is also involved in blood pressure regulation and cleaves a range of substrates involved in different physiological processes. Importantly, it is the functional receptor for severe acute respiratory syndrome (SARS)-coronavirus (CoV)-2 responsible for the 2020, coronavirus infectious disease 2019 (COVID-19) pandemic. Understanding the interaction between SARS-CoV-2 and ACE2 is crucial for the design of therapies to combat this disease. This review provides a comparative analysis of methodologies and findings to describe how structural biology techniques like X-ray crystallography and cryo-electron microscopy have enabled remarkable discoveries into the structure-function relationship of ACE and ACE2. This, in turn, has enabled the development of ACE inhibitors for the treatment of cardiovascular disease and candidate therapies for the treatment of COVID-19. However, despite these advances the function of ACE homologues in non-human organisms is not yet fully understood. ACE homologues have been discovered in the tissues, body fluids and venom of species from diverse lineages and are known to have important functions in fertility, envenoming and insect-host defence mechanisms. We, therefore, further highlight the need for structural insight into insect and venom ACE homologues for the potential development of novel anti-venoms and insecticides.
Keywords: ACE inhibitor; COVID-19; angiotensin converting enzyme 2; metalloproteases.
© 2020 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

