Dienone Compounds: Targets and Pharmacological Responses
- PMID: 33146523
- PMCID: PMC7770821
- DOI: 10.1021/acs.jmedchem.0c00812
Dienone Compounds: Targets and Pharmacological Responses
Abstract
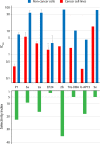
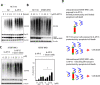
The biological responses to dienone compounds with a 1,5-diaryl-3-oxo-1,4-pentadienyl pharmacophore have been studied extensively. Despite their expected general thiol reactivity, these compounds display considerable degrees of tumor cell selectivity. Here we review in vitro and preclinical studies of dienone compounds including b-AP15, VLX1570, RA-9, RA-190, EF24, HO-3867, and MCB-613. A common property of these compounds is their targeting of the ubiquitin-proteasome system (UPS), known to be essential for the viability of tumor cells. Gene expression profiling experiments have shown induction of responses characteristic of UPS inhibition, and experiments using cellular reporter proteins have shown that proteasome inhibition is associated with cell death. Other mechanisms of action such as reactivation of mutant p53, stimulation of steroid receptor coactivators, and induction of protein cross-linking have also been described. Although unsuitable as biological probes due to widespread reactivity, dienone compounds are cytotoxic to apoptosis-resistant tumor cells and show activity in animal tumor models.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


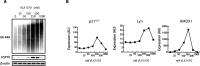


References
-
- Lamb J.; Crawford E. D.; Peck D.; Modell J. W.; Blat I. C.; Wrobel M. J.; Lerner J.; Brunet J. P.; Subramanian A.; Ross K. N.; Reich M.; Hieronymus H.; Wei G.; Armstrong S. A.; Haggarty S. J.; Clemons P. A.; Wei R.; Carr S. A.; Lander E. S.; Golub T. R. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science 2006, 313, 1929–1935. 10.1126/science.1132939. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

