Reconstruction of natural images from responses of primate retinal ganglion cells
- PMID: 33146609
- PMCID: PMC7752138
- DOI: 10.7554/eLife.58516
Reconstruction of natural images from responses of primate retinal ganglion cells
Abstract
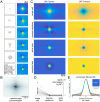
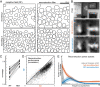
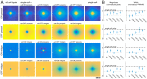
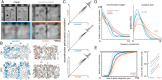
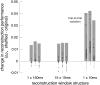
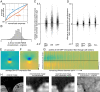
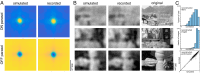
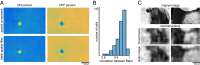
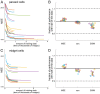
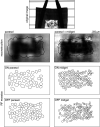
The visual message conveyed by a retinal ganglion cell (RGC) is often summarized by its spatial receptive field, but in principle also depends on the responses of other RGCs and natural image statistics. This possibility was explored by linear reconstruction of natural images from responses of the four numerically-dominant macaque RGC types. Reconstructions were highly consistent across retinas. The optimal reconstruction filter for each RGC - its visual message - reflected natural image statistics, and resembled the receptive field only when nearby, same-type cells were included. ON and OFF cells conveyed largely independent, complementary representations, and parasol and midget cells conveyed distinct features. Correlated activity and nonlinearities had statistically significant but minor effects on reconstruction. Simulated reconstructions, using linear-nonlinear cascade models of RGC light responses that incorporated measured spatial properties and nonlinearities, produced similar results. Spatiotemporal reconstructions exhibited similar spatial properties, suggesting that the results are relevant for natural vision.
Keywords: decoding; macaca fascicularis; natural images; neuroscience; reconstruction; retina; retinal ganglion cells; rhesus macaque.
Plain language summary
Vision begins in the retina, the layer of tissue that lines the back of the eye. Light-sensitive cells called rods and cones absorb incoming light and convert it into electrical signals. They pass these signals to neurons called retinal ganglion cells (RGCs), which convert them into electrical signals called spikes. Spikes from RGCs then travel along the optic nerve to the brain. They are the only source of visual information that the brain receives. From this information, the brain constructs our entire visual world. The primate retina contains roughly 20 types of RGCs. Each encodes a different visual feature, such as the presence of bright spots of a certain size, or information about texture and movement. But exactly what input each RGC sends to the brain, and how the brain uses this information, is unclear. Brackbill et al. set out to answer these questions by measuring and analyzing the electrical activity in isolated retinas from macaque monkeys. Studying the macaque retina was important because the primate visual system differs from that of other species in several ways. These include the numbers and types of RGCs present in the retina. These primates are also similar to humans in their high-resolution central vision and trichromatic color vision. Using electrode arrays to monitor hundreds of RGCs at the same time, Brackbill et al. recorded the responses of macaque retinas to real-life images of landscapes, objects, animals or people. Based on these recordings, plus existing knowledge about RGC responses, Brackbill et al. then attempted to reconstruct the original images using just the electrical activity recorded. The resulting reconstructions were similar across all retinas tested. Moreover, they showed a striking resemblance to the original images. These results made it possible to comprehend how the light-response properties of each cell represent visual information that can be used by the brain. Understanding how macaque retinas work in natural conditions is critical to decoding how our own retinas process and convey information. A better knowledge of how the brain uses this input to generate images could ultimately make it possible to design artificial retinas to restore vision in patients with certain forms of blindness.
© 2020, Brackbill et al.
Conflict of interest statement
NB, CR, AK, NS, AS, AL, EC No competing interests declared
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

