R-loops at centromeric chromatin contribute to defects in kinetochore integrity and chromosomal instability in budding yeast
- PMID: 33147102
- PMCID: PMC8098821
- DOI: 10.1091/mbc.E20-06-0379
R-loops at centromeric chromatin contribute to defects in kinetochore integrity and chromosomal instability in budding yeast
Abstract
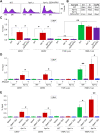
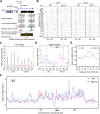
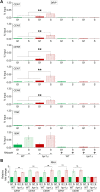
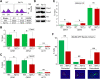
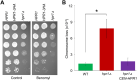
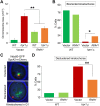
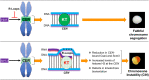
R-loops, the byproduct of DNA-RNA hybridization and the displaced single-stranded DNA (ssDNA), have been identified in bacteria, yeasts, and other eukaryotic organisms. The persistent presence of R-loops contributes to defects in DNA replication and repair, gene expression, and genomic integrity. R-loops have not been detected at centromeric (CEN) chromatin in wild-type budding yeast. Here we used an hpr1∆ strain that accumulates R-loops to investigate the consequences of R-loops at CEN chromatin and chromosome segregation. We show that Hpr1 interacts with the CEN-histone H3 variant, Cse4, and prevents the accumulation of R-loops at CEN chromatin for chromosomal stability. DNA-RNA immunoprecipitation (DRIP) analysis showed an accumulation of R-loops at CEN chromatin that was reduced by overexpression of RNH1 in hpr1∆ strains. Increased levels of ssDNA, reduced levels of Cse4 and its assembly factor Scm3, and mislocalization of histone H3 at CEN chromatin were observed in hpr1∆ strains. We determined that accumulation of R-loops at CEN chromatin contributes to defects in kinetochore biorientation and chromosomal instability (CIN) and these phenotypes are suppressed by RNH1 overexpression in hpr1∆ strains. In summary, our studies provide mechanistic insights into how accumulation of R-loops at CEN contributes to defects in kinetochore integrity and CIN.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

