Loss of hepatocyte cell division leads to liver inflammation and fibrosis
- PMID: 33147210
- PMCID: PMC7641358
- DOI: 10.1371/journal.pgen.1009084
Loss of hepatocyte cell division leads to liver inflammation and fibrosis
Abstract
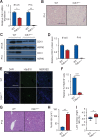
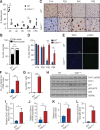
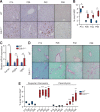
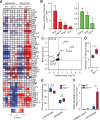
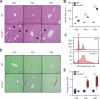
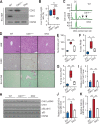
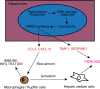
The liver possesses a remarkable regenerative capacity based partly on the ability of hepatocytes to re-enter the cell cycle and divide to replace damaged cells. This capability is substantially reduced upon chronic damage, but it is not clear if this is a cause or consequence of liver disease. Here, we investigate whether blocking hepatocyte division using two different mouse models affects physiology as well as clinical liver manifestations like fibrosis and inflammation. We find that in P14 Cdk1Liv-/- mice, where the division of hepatocytes is abolished, polyploidy, DNA damage, and increased p53 signaling are prevalent. Cdk1Liv-/- mice display classical markers of liver damage two weeks after birth, including elevated ALT, ALP, and bilirubin levels, despite the lack of exogenous liver injury. Inflammation was further studied using cytokine arrays, unveiling elevated levels of CCL2, TIMP1, CXCL10, and IL1-Rn in Cdk1Liv-/- liver, which resulted in increased numbers of monocytes. Ablation of CDK2-dependent DNA re-replication and polyploidy in Cdk1Liv-/- mice reversed most of these phenotypes. Overall, our data indicate that blocking hepatocyte division induces biological processes driving the onset of the disease phenotype. It suggests that the decrease in hepatocyte division observed in liver disease may not only be a consequence of fibrosis and inflammation, but also a pathological cue.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Similar articles
-
c-Met confers protection against chronic liver tissue damage and fibrosis progression after bile duct ligation in mice.Gastroenterology. 2009 Jul;137(1):297-308, 308.e1-4. doi: 10.1053/j.gastro.2009.01.068. Epub 2009 Feb 5. Gastroenterology. 2009. PMID: 19208365
-
Loss of cellular FLICE-inhibitory protein promotes acute cholestatic liver injury and inflammation from bile duct ligation.Am J Physiol Gastrointest Liver Physiol. 2018 Mar 1;314(3):G319-G333. doi: 10.1152/ajpgi.00097.2017. Epub 2017 Nov 30. Am J Physiol Gastrointest Liver Physiol. 2018. PMID: 29191940
-
Essential role of suppressor of cytokine signaling 1 (SOCS1) in hepatocytes and macrophages in the regulation of liver fibrosis.Cytokine. 2019 Dec;124:154501. doi: 10.1016/j.cyto.2018.07.032. Epub 2018 Aug 8. Cytokine. 2019. PMID: 30097285
-
Hepatocyte-specific TAK1 deficiency drives RIPK1 kinase-dependent inflammation to promote liver fibrosis and hepatocellular carcinoma.Proc Natl Acad Sci U S A. 2020 Jun 23;117(25):14231-14242. doi: 10.1073/pnas.2005353117. Epub 2020 Jun 8. Proc Natl Acad Sci U S A. 2020. PMID: 32513687 Free PMC article.
-
Crosstalk network among multiple inflammatory mediators in liver fibrosis.World J Gastroenterol. 2019 Sep 7;25(33):4835-4849. doi: 10.3748/wjg.v25.i33.4835. World J Gastroenterol. 2019. PMID: 31543677 Free PMC article. Review.
Cited by
-
Fat storage-inducing transmembrane proteins: beyond mediating lipid droplet formation.Cell Mol Biol Lett. 2022 Nov 8;27(1):98. doi: 10.1186/s11658-022-00391-z. Cell Mol Biol Lett. 2022. PMID: 36348306 Free PMC article. Review.
-
Assessment of Drug-Induced Liver Injury through Cell Morphology and Gene Expression Analysis.Chem Res Toxicol. 2023 Sep 18;36(9):1456-1470. doi: 10.1021/acs.chemrestox.2c00381. Epub 2023 Aug 31. Chem Res Toxicol. 2023. PMID: 37652439 Free PMC article.
-
Assessment of Hepatocyte Ploidy by Flow Cytometry.Methods Mol Biol. 2022;2544:171-181. doi: 10.1007/978-1-0716-2557-6_12. Methods Mol Biol. 2022. PMID: 36125718
-
A network-based transcriptomic landscape of HepG2 cells uncovering causal gene-cytotoxicity interactions underlying drug-induced liver injury.Toxicol Sci. 2024 Feb 28;198(1):14-30. doi: 10.1093/toxsci/kfad121. Toxicol Sci. 2024. PMID: 38015832 Free PMC article.
-
Effect of Hepatic Pathology on Liver Regeneration: The Main Metabolic Mechanisms Causing Impaired Hepatic Regeneration.Int J Mol Sci. 2023 May 23;24(11):9112. doi: 10.3390/ijms24119112. Int J Mol Sci. 2023. PMID: 37298064 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

