Bone marrow microenvironments that contribute to patient outcomes in newly diagnosed multiple myeloma: A cohort study of patients in the Total Therapy clinical trials
- PMID: 33147277
- PMCID: PMC7641353
- DOI: 10.1371/journal.pmed.1003323
Bone marrow microenvironments that contribute to patient outcomes in newly diagnosed multiple myeloma: A cohort study of patients in the Total Therapy clinical trials
Erratum in
-
Correction: Bone marrow microenvironments that contribute to patient outcomes in newly diagnosed multiple myeloma: A cohort study of patients in the Total Therapy clinical trials.PLoS Med. 2021 Sep 8;18(9):e1003774. doi: 10.1371/journal.pmed.1003774. eCollection 2021 Sep. PLoS Med. 2021. PMID: 34495968 Free PMC article.
Abstract
Background: The tumor microenvironment (TME) is increasingly appreciated as an important determinant of cancer outcome, including in multiple myeloma (MM). However, most myeloma microenvironment studies have been based on bone marrow (BM) aspirates, which often do not fully reflect the cellular content of BM tissue itself. To address this limitation in myeloma research, we systematically characterized the whole bone marrow (WBM) microenvironment during premalignant, baseline, on treatment, and post-treatment phases.
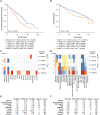
Methods and findings: Between 2004 and 2019, 998 BM samples were taken from 436 patients with newly diagnosed MM (NDMM) at the University of Arkansas for Medical Sciences in Little Rock, Arkansas, United States of America. These patients were 61% male and 39% female, 89% White, 8% Black, and 3% other/refused, with a mean age of 58 years. Using WBM and matched cluster of differentiation (CD)138-selected tumor gene expression to control for tumor burden, we identified a subgroup of patients with an adverse TME associated with 17 fewer months of progression-free survival (PFS) (95% confidence interval [CI] 5-29, 49-69 versus 70-82 months, χ2 p = 0.001) and 15 fewer months of overall survival (OS; 95% CI -1 to 31, 92-120 versus 113-129 months, χ2 p = 0.036). Using immunohistochemistry-validated computational tools that identify distinct cell types from bulk gene expression, we showed that the adverse outcome was correlated with elevated CD8+ T cell and reduced granulocytic cell proportions. This microenvironment develops during the progression of premalignant to malignant disease and becomes less prevalent after therapy, in which it is associated with improved outcomes. In patients with quantified International Staging System (ISS) stage and 70-gene Prognostic Risk Score (GEP-70) scores, taking the microenvironment into consideration would have identified an additional 40 out of 290 patients (14%, premutation p = 0.001) with significantly worse outcomes (PFS, 95% CI 6-36, 49-73 versus 74-90 months) who were not identified by existing clinical (ISS stage III) and tumor (GEP-70) criteria as high risk. The main limitations of this study are that it relies on computationally identified cell types and that patients were treated with thalidomide rather than current therapies.
Conclusions: In this study, we observe that granulocyte signatures in the MM TME contribute to a more accurate prognosis. This implies that future researchers and clinicians treating patients should quantify TME components, in particular monocytes and granulocytes, which are often ignored in microenvironment studies.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: SAD, MM, MHY, FS, DJR, YR, KS, WBC, BF, AF, KN, APD, and AVR declare employment and equity ownership for Bristol Myers Squibb. JG declares previous employment at Celgene Corporation. SSC declares previous employment and equity ownership at Celgene Corporation. AR, PF, DVA, AS, CA, MB, FVR, MZ, NP, AH, and GJM declare no competing financial interests. FD declares consultancy for AbbVie; consultancy and membership on an entity’s Board of Directors or advisory committee for Amgen, Roche, and Takeda; consultancy, honoraria, membership on an entity’s Board of Directors or advisory committee, and research funding from Celgene Corporation, a wholly-owned subsidiary of Bristol Myers Squibb; consultancy and honoraria from Janssen; honoraria from TRM Oncology. RZO declares honoraria from Abbott Laboratories, Amgen, Array BioPharma, BioTheryX, Bristol Myers Squibb, Celgene Corporation a wholly-owned subsidiary of Bristol Myers Squibb, Cephalon, Inc., Forma Therapeutics, Genentech, Inc., Incyte, Janssen-Cilag, Janssen R&D, Millennium Pharmaceuticals, and Novartis; research funding from Amgen, Array BioPharma, Bristol Myers Squibb, Celgene Corporation a wholly-owned subsidiary of Bristol Myers Squibb, Janssen-Cilag, Janssen R&D, Millennium Pharmaceuticals, Onyx Pharmaceuticals, and Spectrum Pharmaceuticals. MVD declares membership on an entity’s Board of Directors or advisory committee for Amgen, Bristol Myers Squib, Celgene Corporation a wholly owned subsidiary of Bristol Myers Squibb, Janssen, Lava Therapeutics, and Roche. BB declares consultancy and research funding from Celgene Corporation a wholly owned subsidiary of Bristol Myers Squibb and Millennium Pharmaceuticals; travel funding from Comtecmed, Dana-Farber Cancer Institute, European School of Hematology, International Workshop on Waldenström Macroglobulinemia, and The Multiple Myeloma Research Foundation; patents and royalties from Myeloma Health LLC. MWBT declares employment at Celgene Corporation, a Bristol Myers Squibb Company (Spain) and equity ownership for Bristol Myers Squibb Company. RMH declares employment and equity ownership for Bristol Myers Squibb; membership on an entity’s Board of Directors or advisory committee for Adaptive Biotechnologies and NanoString Technologies; consultancy at Frazier Healthcare Partners. BAW declares research funding from Celgene Corporation a wholly owned subsidiary of Bristol Myers Squibb.
Figures





References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

