Identification of Required Host Factors for SARS-CoV-2 Infection in Human Cells
- PMID: 33147445
- PMCID: PMC7584921
- DOI: 10.1016/j.cell.2020.10.030
Identification of Required Host Factors for SARS-CoV-2 Infection in Human Cells
Abstract
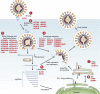
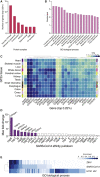
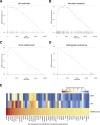
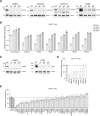
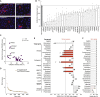
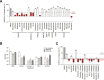
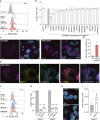
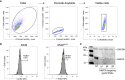
To better understand host-virus genetic dependencies and find potential therapeutic targets for COVID-19, we performed a genome-scale CRISPR loss-of-function screen to identify host factors required for SARS-CoV-2 viral infection of human alveolar epithelial cells. Top-ranked genes cluster into distinct pathways, including the vacuolar ATPase proton pump, Retromer, and Commander complexes. We validate these gene targets using several orthogonal methods such as CRISPR knockout, RNA interference knockdown, and small-molecule inhibitors. Using single-cell RNA-sequencing, we identify shared transcriptional changes in cholesterol biosynthesis upon loss of top-ranked genes. In addition, given the key role of the ACE2 receptor in the early stages of viral entry, we show that loss of RAB7A reduces viral entry by sequestering the ACE2 receptor inside cells. Overall, this work provides a genome-scale, quantitative resource of the impact of the loss of each host gene on fitness/response to viral infection.
Keywords: COVID-19; CRISPR; Cas9; ECCITE-seq; SARS-CoV-2; cholesterol; endosome; genome-wide screen; human lung; loss of function.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests N.E.S. is an advisor to Vertex. T.L. is a member of the scientific advisory boards of Goldfinch Bio and, with equity, Variant Bio. The New York Genome Center and New York University have applied for patents relating to the work in this article.
Figures





Comment in
-
Host genetics of coronavirus infection.Nat Rev Genet. 2021 Jan;22(1):1. doi: 10.1038/s41576-020-00310-y. Nat Rev Genet. 2021. PMID: 33235360 Free PMC article.
-
Cellular basis for SARS-CoV-2 infection.Nat Rev Mol Cell Biol. 2021 Jan;22(1):2. doi: 10.1038/s41580-020-00319-5. Nat Rev Mol Cell Biol. 2021. PMID: 33262537 Free PMC article.
-
A Crisp(r) New Perspective on SARS-CoV-2 Biology.Cell. 2021 Jan 7;184(1):15-17. doi: 10.1016/j.cell.2020.12.003. Epub 2020 Dec 17. Cell. 2021. PMID: 33338422 Free PMC article.
-
The COVID-19 susceptibility of cancer patients might due to the high expression of SARS-CoV-2 required host factors.J Infect. 2022 Mar;84(3):418-467. doi: 10.1016/j.jinf.2021.12.011. Epub 2021 Dec 22. J Infect. 2022. PMID: 34953912 Free PMC article. No abstract available.
References
-
- Aguet F., Barbeira A.N., Bonazzola R., Brown A., Castel S.E., Jo B., Kasela S., Kim-Hellmuth S., Liang Y., Oliva M., et al. The GTEx Consortium atlas of genetic regulatory effects across human tissues. bioRxiv. 2019 doi: 10.1101/787903. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

