Human and Mouse Transcriptome Profiling Identifies Cross-Species Homology in Pulmonary and Lymph Node Mononuclear Phagocytes
- PMID: 33147458
- PMCID: PMC7673261
- DOI: 10.1016/j.celrep.2020.108337
Human and Mouse Transcriptome Profiling Identifies Cross-Species Homology in Pulmonary and Lymph Node Mononuclear Phagocytes
Abstract
The mononuclear phagocyte (MP) system consists of macrophages, monocytes, and dendritic cells (DCs). MP subtypes play distinct functional roles in steady-state and inflammatory conditions. Although murine MPs are well characterized, their pulmonary and lymph node (LN) human homologs remain poorly understood. To address this gap, we have created a gene expression compendium across 24 distinct human and murine lung and LN MPs, along with human blood and murine spleen MPs, to serve as validation datasets. In-depth RNA sequencing identifies corresponding human-mouse MP subtypes and determines marker genes shared and divergent across species. Unexpectedly, only 13%-23% of the top 1,000 marker genes (i.e., genes not shared across species-specific MP subtypes) overlap in corresponding human-mouse MP counterparts. Lastly, CD88 in both species helps distinguish monocytes/macrophages from DCs. Our cross-species expression compendium serves as a resource for future translational studies to investigate beforehand whether pursuing specific MP subtypes or genes will prove fruitful.
Keywords: dendritic cells; human; lung; macrophages; monocytes; mononuclear phagocytes; pulmonary.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures

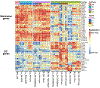


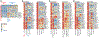

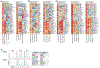
References
-
- Alferink J, Lieberam I, Reindl W, Behrens A, Weiss S, Hüser N, Gerauer K, Ross R, Reske-Kunz AB, Ahmad-Nejad P, et al. (2003). Compartmentalized production of CCL17 in vivo: strong inducibility in peripheral dendritic cells contrasts selective absence from the spleen. J. Exp. Med 197, 585–599. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

