Interferon Regulatory Factors IRF1 and IRF7 Directly Regulate Gene Expression in Bats in Response to Viral Infection
- PMID: 33147460
- PMCID: PMC8755441
- DOI: 10.1016/j.celrep.2020.108345
Interferon Regulatory Factors IRF1 and IRF7 Directly Regulate Gene Expression in Bats in Response to Viral Infection
Abstract
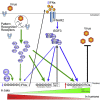
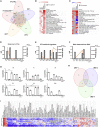
Bat cells and tissue have elevated basal expression levels of antiviral genes commonly associated with interferon alpha (IFNα) signaling. Here, we show Interferon Regulatory Factor 1 (IRF1), 3, and 7 levels are elevated in most bat tissues and that, basally, IRFs contribute to the expression of type I IFN ligands and high expression of interferon regulated genes (IRGs). CRISPR knockout (KO) of IRF 1/3/7 in cells reveals distinct subsets of genes affected by each IRF in an IFN-ligand signaling-dependent and largely independent manner. As the master regulators of innate immunity, the IRFs control the kinetics and maintenance of the IRG response and play essential roles in response to influenza A virus (IAV), herpes simplex virus 1 (HSV-1), Melaka virus/Pteropine orthoreovirus 3 Melaka (PRV3M), and Middle East respiratory syndrome-related coronavirus (MERS-CoV) infection. With its differential expression in bats compared to that in humans, this highlights a critical role for basal IRF expression in viral responses and potentially immune cell development in bats with relevance for IRF function in human biology.
Keywords: IAV; IRF; IRG; Interferon Regulatory Factor; MERS; PRV3M; bat; innate immunity; interferon; virus.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests We declare there is no conflict of interest.
Figures




References
-
- Austad S.N., Fischer K.E. Mammalian aging, metabolism, and ecology: Evidence from the bats and marsupials. J. Gerontol. 1991;46:B47–B53. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

