The Domestic Environment and the Lung Mycobiome
- PMID: 33147738
- PMCID: PMC7693370
- DOI: 10.3390/microorganisms8111717
The Domestic Environment and the Lung Mycobiome
Abstract
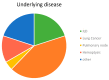
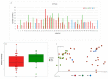
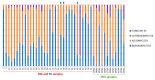
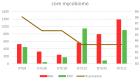
This study analyzes the relationship between the mycobiome of the Lower Respiratory Tract (LRT) and the fungi in the domestic environment. Samples studied consisted of Broncho-Alveolar Lavage (BAL) from 45 patients who underwent bronchoscopy for different diagnostic purposes, and dust and air from the houses (ENV) of 20 of them (44.4%). Additionally, five bronchoscopes (BS) were also analyzed and negative controls were included for every procedure. All samples were processed for DNA extraction and cultures, which were performed in Sabouraud Dextrose and Potato Dextrose Agar. The fungal Internal Transcribed Spacer (ITS2) was sequenced by the Solexa/Illumina system and sequences were analyzed by QIIME 1.8.0 and compared with the UNITE Database for identification. The similarity between the two fungal communities (BAL and ENV) for a specific patient was assessed via the percentage of coincidence in the detection of specific operational taxonomic units (OTUs), and about 75% of co-occurrence was detected between the mycobiome of the LRT and the houses. Cultures confirmed the presence of the core mycobiome species. However, the low rate of isolation from BAL suggests that most of its mycobiome corresponds to non-culturable cells. This likely depends on the patient's immune system activity and inflammatory status.
Keywords: bronco-alveolar lavage; fungi; house dust; lower respiratory tract; mycobiome; mycobiota.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Figures

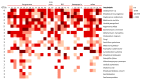
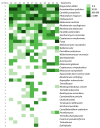

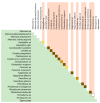
References
-
- Thomas S., Izard J., Walsh E., Batich K., Chongsathidkiet P., Clarke G., Sela D.A., Muller A.J., Mullin J.M., Albert K., et al. The Host Microbiome Regulates and Maintains Human Health: A Primer and Perspective for Non-Microbiologists. Cancer Res. 2017;77:1783–1812. doi: 10.1158/0008-5472.CAN-16-2929. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

