Multiclonal complexity of pediatric acute lymphoblastic leukemia and the prognostic relevance of subclonal mutations
- PMID: 33147938
- PMCID: PMC8634184
- DOI: 10.3324/haematol.2020.259226
Multiclonal complexity of pediatric acute lymphoblastic leukemia and the prognostic relevance of subclonal mutations
Abstract
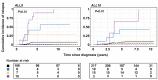
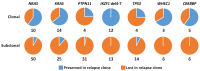
Genomic studies of pediatric acute lymphoblastic leukemia (ALL) have shown remarkable heterogeneity in initial diagnosis, with multiple (sub)clones harboring lesions in relapse-associated genes. However, the clinical relevance of these subclonal alterations remains unclear. We assessed the clinical relevance and prognostic value of subclonal alterations in the relapse-associated genes IKZF1, CREBBP, KRAS, NRAS, PTPN11, TP53, NT5C2, and WHSC1 in 503 ALL cases. Using Molecular Inversion Probe sequencing and breakpoint-spanning PCR we reliably detected alterations below 1% allele frequency. We identified 660 genomic alterations in 285 diagnosis samples of which 495 (75%) were subclonal. RAS pathway mutations were common, particularly in minor subclones, and comparisons between RAS hotspot mutations revealed differences in their capacity to drive clonal expansion in ALL. We did not find an association of subclonal alterations with unfavorable outcome. Particularly for IKZF1, an established prognostic marker in ALL, all clonal but none of the subclonal alterations were preserved at relapse. We conclude that, for the genes tested, there is no basis to consider subclonal alterations detected at diagnosis for risk group stratification of ALL treatment.
Figures
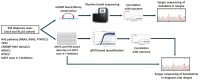
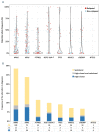
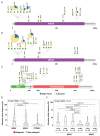
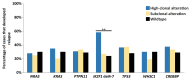
References
-
- Pieters R, de Groot-Kruseman Hd, Velden V, et al. Successful therapy reduction and intensification for childhood acute lymphoblastic leukemia based on minimal residual disease monitoring: study ALL10 from the Dutch Childhood Oncology Group. J Clin Oncol. 2016;34(22):2591-2601. - PubMed
-
- Anderson K, Lutz C, van Delft FW, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469(7330):356-361. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

