Unraveling Reversible DNA Cross-Links with a Biological Machine
- PMID: 33147957
- PMCID: PMC7700721
- DOI: 10.1021/acs.chemrestox.0c00413
Unraveling Reversible DNA Cross-Links with a Biological Machine
Abstract
The reversible generation and capture of certain electrophilic quinone methide intermediates support dynamic reactions with DNA that allow for migration and transfer of alkylation and cross-linking. This reversibility also expands the possible consequences that can be envisioned when confronted by DNA repair processes and biological machines. To begin testing the response to such an encounter, quinone methide-based modification of DNA has now been challenged with a helicase (T7 bacteriophage gene protein four, T7gp4) that promotes 5' to 3' translocation and unwinding. This model protein was selected based on its widespread application, well characterized mechanism and detailed structural information. Little over one-half of the cross-linking generated by a bisfunctional quinone methide remained stable to T7gp4 and did not suppress its activity. The helicase likely avoids the topological block generated by this fraction of cross-linking by its ability to shift from single- to double-stranded translocation. The remaining fraction of cross-linking was destroyed during T7gp4 catalysis. Thus, this helicase is chemically competent to promote release of the quinone methide from DNA. The ability of T7gp4 to act as a Brownian ratchet for unwinding DNA may block recapture of the QM intermediate by DNA during its transient release from a donor strand. Most surprisingly, T7gp4 releases the quinone methide from both the translocating strand that passes through its central channel and the excluded strand that was typically unaffected by other lesions. The ability of T7gp4 to reverse the cross-link formed by the quinone methide does not extend to that formed irreversibly by the nitrogen mustard mechlorethamine.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




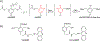
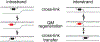
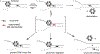
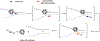
Similar articles
-
Conjugation of a hairpin pyrrole-imidazole polyamide to a quinone methide for control of DNA cross-linking.Bioconjug Chem. 2004 Jul-Aug;15(4):915-22. doi: 10.1021/bc049941h. Bioconjug Chem. 2004. PMID: 15264882
-
Directing Quinone Methide-Dependent Alkylation and Cross-Linking of Nucleic Acids with Quaternary Amines.Bioconjug Chem. 2020 May 20;31(5):1486-1496. doi: 10.1021/acs.bioconjchem.0c00166. Epub 2020 Apr 23. Bioconjug Chem. 2020. PMID: 32298588 Free PMC article.
-
Quinone methide derivatives: important intermediates to DNA alkylating and DNA cross-linking actions.Curr Med Chem. 2005;12(24):2893-913. doi: 10.2174/092986705774454724. Curr Med Chem. 2005. PMID: 16305478 Review.
-
Inducible alkylation of DNA by a quinone methide-peptide nucleic acid conjugate.Biochemistry. 2012 Feb 7;51(5):1020-7. doi: 10.1021/bi201492b. Epub 2012 Jan 24. Biochemistry. 2012. PMID: 22243337 Free PMC article.
-
The unexplored potential of quinone methides in chemical biology.Bioorg Med Chem. 2019 Jun 15;27(12):2298-2305. doi: 10.1016/j.bmc.2019.04.001. Epub 2019 Apr 2. Bioorg Med Chem. 2019. PMID: 30955994 Review.
References
-
- Geacintov NE, Cosman M, Hingerty BE, Amin S, Broyde S, and Patel DJ (1997) NMR solution structures of stereoisomeric covalent polycyclic aromatic carcinogen-DNA adducts: principles, patterns and diversity. Chem. Res. Toxicol 10, 111–146. - PubMed
-
- Guengerich FP (2006) Interactions of carcinogen-bound DNA with individual DNA polymerases. Chem. Rev 106, 420–452. - PubMed
-
- Plastaras JP, Dedon PC, and Marnett LJ (2002) Effects of DNA structure on oxopropenylation by the endogenous mutagens malondialdehyde and base propenal. Biochemistry 41, 5033–5042. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

