Polymerization of Bacillus subtilis MreB on a lipid membrane reveals lateral co-polymerization of MreB paralogs and strong effects of cations on filament formation
- PMID: 33148162
- PMCID: PMC7641798
- DOI: 10.1186/s12860-020-00319-5
Polymerization of Bacillus subtilis MreB on a lipid membrane reveals lateral co-polymerization of MreB paralogs and strong effects of cations on filament formation
Abstract
Background: MreB is a bacterial ortholog of actin and forms mobile filaments underneath the cell membrane, perpendicular to the long axis of the cell, which play a crucial role for cell shape maintenance. We wished to visualize Bacillus subtilis MreB in vitro and therefore established a protocol to obtain monomeric protein, which could be polymerized on a planar membrane system, or associated with large membrane vesicles.
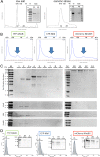
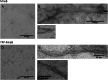
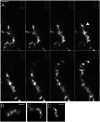
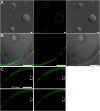
Results: Using a planar membrane system and electron microscopy, we show that Bacillus subtilis MreB forms bundles of filaments, which can branch and fuse, with an average width of 70 nm. Fluorescence microscopy of non-polymerized YFP-MreB, CFP-Mbl and mCherry-MreBH proteins showed uniform binding to the membrane, suggesting that 2D diffusion along the membrane could facilitate filament formation. After addition of divalent magnesium and calcium ions, all three proteins formed highly disordered sheets of filaments that could split up or merge, such that at high protein concentration, MreB and its paralogs generated a network of filaments extending away from the membrane. Filament formation was positively affected by divalent ions and negatively by monovalent ions. YFP-MreB or CFP-Mbl also formed filaments between two adjacent membranes, which frequently has a curved appearance. New MreB, Mbl or MreBH monomers could add to the lateral side of preexisting filaments, and MreB paralogs co-polymerized, indicating direct lateral interaction between MreB paralogs.
Conclusions: Our data show that B. subtilis MreB paralogs do not easily form ordered filaments in vitro, possibly due to extensive lateral contacts, but can co-polymerise. Monomeric MreB, Mbl and MreBH uniformly bind to a membrane, and form irregular and frequently split up filamentous structures, facilitated by the addition of divalent ions, and counteracted by monovalent ions, suggesting that intracellular potassium levels may be one important factor to counteract extensive filament formation and filament splitting in vivo.
Conflict of interest statement
The authors declare that no competing financial or scientific interests exist.
Figures



Similar articles
-
Bacillus subtilis MreB orthologs self-organize into filamentous structures underneath the cell membrane in a heterologous cell system.PLoS One. 2011;6(11):e27035. doi: 10.1371/journal.pone.0027035. Epub 2011 Nov 1. PLoS One. 2011. PMID: 22069484 Free PMC article.
-
Bacillus subtilis MreB paralogues have different filament architectures and lead to shape remodelling of a heterologous cell system.Mol Microbiol. 2010 Dec;78(5):1145-58. doi: 10.1111/j.1365-2958.2010.07395.x. Epub 2010 Oct 8. Mol Microbiol. 2010. PMID: 21091501
-
Translation elongation factor EF-Tu modulates filament formation of actin-like MreB protein in vitro.J Mol Biol. 2015 Apr 24;427(8):1715-27. doi: 10.1016/j.jmb.2015.01.025. Epub 2015 Feb 10. J Mol Biol. 2015. PMID: 25676310
-
The actin-like MreB proteins in Bacillus subtilis: a new turn.Front Biosci (Schol Ed). 2012 Jun 1;4(4):1582-606. doi: 10.2741/s354. Front Biosci (Schol Ed). 2012. PMID: 22652894 Review.
-
Increasing complexity of the bacterial cytoskeleton.Curr Opin Cell Biol. 2005 Feb;17(1):75-81. doi: 10.1016/j.ceb.2004.11.002. Curr Opin Cell Biol. 2005. PMID: 15661522 Review.
Cited by
-
On the role of nucleotides and lipids in the polymerization of the actin homolog MreB from a Gram-positive bacterium.Elife. 2023 Oct 11;12:e84505. doi: 10.7554/eLife.84505. Elife. 2023. PMID: 37818717 Free PMC article.
-
Adaptation of Bacillus subtilis MreB Filaments to Osmotic Stress Depends on Influx of Potassium Ions.Microorganisms. 2024 Jun 27;12(7):1309. doi: 10.3390/microorganisms12071309. Microorganisms. 2024. PMID: 39065078 Free PMC article.
-
Molecular motor tug-of-war regulates elongasome cell wall synthesis dynamics in Bacillus subtilis.Nat Commun. 2024 Jun 26;15(1):5411. doi: 10.1038/s41467-024-49785-x. Nat Commun. 2024. PMID: 38926336 Free PMC article.
-
Scattering-based Light Microscopy: From Metal Nanoparticles to Single Proteins.Chem Rev. 2021 Oct 13;121(19):11937-11970. doi: 10.1021/acs.chemrev.1c00271. Epub 2021 Sep 29. Chem Rev. 2021. PMID: 34587448 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

