Eukaryotic translation initiation factors as promising targets in cancer therapy
- PMID: 33148274
- PMCID: PMC7640403
- DOI: 10.1186/s12964-020-00607-9
Eukaryotic translation initiation factors as promising targets in cancer therapy
Abstract
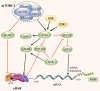
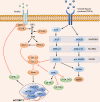
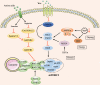
The regulation of the translation of messenger RNA (mRNA) in eukaryotic cells is critical for gene expression, and occurs principally at the initiation phase which is mainly regulated by eukaryotic initiation factors (eIFs). eIFs are fundamental for the translation of mRNA and as such act as the primary targets of several signaling pathways to regulate gene expression. Mis-regulated mRNA expression is a common feature of tumorigenesis and the abnormal activity of eIF complexes triggered by upstream signaling pathways is detected in many tumors, leading to the selective translation of mRNA encoding proteins involved in tumorigenesis, metastasis, or resistance to anti-cancer drugs, and making eIFs a promising therapeutic target for various types of cancers. Here, we briefly outline our current understanding of the biology of eIFs, mainly focusing on the effects of several signaling pathways upon their functions and discuss their contributions to the initiation and progression of tumor growth. An overview of the progress in developing agents targeting the components of translation machinery for cancer treatment is also provided. Video abstract.
Keywords: Cancer; MAPK; PI3K/Akt; eIF; mRNA translation; mTOR.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

Similar articles
-
Potential therapeutic targets of eukaryotic translation initiation factors in tumor therapy.Eur J Med Chem. 2025 Jul 5;291:117638. doi: 10.1016/j.ejmech.2025.117638. Epub 2025 Apr 14. Eur J Med Chem. 2025. PMID: 40273663 Review.
-
Eukaryotic translation initiation factors and cancer.Tumour Biol. 2017 Jun;39(6):1010428317709805. doi: 10.1177/1010428317709805. Tumour Biol. 2017. PMID: 28653885
-
Translational control gone awry: a new mechanism of tumorigenesis and novel targets of cancer treatments.Biosci Rep. 2011 Feb;31(1):1-15. doi: 10.1042/BSR20100077. Biosci Rep. 2011. PMID: 20964625 Review.
-
Eukaryotic translation initiation factors in cancer development and progression.Cancer Lett. 2013 Oct 28;340(1):9-21. doi: 10.1016/j.canlet.2013.06.019. Epub 2013 Jul 2. Cancer Lett. 2013. PMID: 23830805 Review.
-
Translation initiation in cancer at a glance.J Cell Sci. 2021 Jan 13;134(1):jcs248476. doi: 10.1242/jcs.248476. J Cell Sci. 2021. PMID: 33441326 Review.
Cited by
-
eIF6 Promotes Gastric Cancer Proliferation and Invasion by Regulating Cell Cycle.Dig Dis Sci. 2024 Sep;69(9):3249-3260. doi: 10.1007/s10620-024-08464-z. Epub 2024 Jul 10. Dig Dis Sci. 2024. PMID: 38987443 Free PMC article.
-
Long-acting Erwinia chrysanthemi, Pegcrisantaspase, induces alternate amino acid biosynthetic pathways in a preclinical model of pancreatic ductal adenocarcinoma.Cancer Metab. 2024 Jun 30;12(1):19. doi: 10.1186/s40170-024-00346-2. Cancer Metab. 2024. PMID: 38951899 Free PMC article.
-
Anti-Growth, Anti-Angiogenic, and Pro-Apoptotic Effects by CX-4945, an Inhibitor of Casein Kinase 2, on HuCCT-1 Human Cholangiocarcinoma Cells via Control of Caspase-9/3, DR-4, STAT-3/STAT-5, Mcl-1, eIF-2α, and HIF-1α.Int J Mol Sci. 2022 Jun 6;23(11):6353. doi: 10.3390/ijms23116353. Int J Mol Sci. 2022. PMID: 35683032 Free PMC article.
-
Targeting astrocytes with in vivo gene addition: Can it rescue loss of brain myelin?Mol Ther. 2024 Jun 5;32(6):1602-1603. doi: 10.1016/j.ymthe.2024.05.010. Epub 2024 May 21. Mol Ther. 2024. PMID: 38776907 Free PMC article. No abstract available.
-
Comparative phytochemistry of flavaglines (= rocaglamides), a group of highly bioactive flavolignans from Aglaia species (Meliaceae).Phytochem Rev. 2022;21(3):725-764. doi: 10.1007/s11101-021-09761-5. Epub 2021 Jun 4. Phytochem Rev. 2022. PMID: 34104125 Free PMC article. Review.
References
-
- Ali MU, Ur Rahman MS, Jia Z, Jiang C. Eukaryotic translation initiation factors and cancer. Tumor Biol. 2017;39:1010428317709805. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

