SHERLOCK and DETECTR: CRISPR-Cas Systems as Potential Rapid Diagnostic Tools for Emerging Infectious Diseases
- PMID: 33148705
- PMCID: PMC8106734
- DOI: 10.1128/JCM.00745-20
SHERLOCK and DETECTR: CRISPR-Cas Systems as Potential Rapid Diagnostic Tools for Emerging Infectious Diseases
Retraction in
-
Retraction for Mustafa and Makhawi, "SHERLOCK and DETECTR: CRISPR-Cas Systems as Potential Rapid Diagnostic Tools for Emerging Infectious Diseases".J Clin Microbiol. 2024 Jan 17;62(1):e0152123. doi: 10.1128/jcm.01521-23. Epub 2023 Dec 15. J Clin Microbiol. 2024. PMID: 38099671 Free PMC article. No abstract available.
Abstract
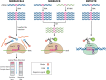
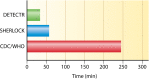
Infectious diseases are one of the most intimidating threats to human race, responsible for an immense burden of disabilities and deaths. Rapid diagnosis and treatment of infectious diseases offers a better understanding of their pathogenesis. According to the World Health Organization, the ideal approach for detecting foreign pathogens should be rapid, specific, sensitive, instrument-free, and cost-effective. Nucleic acid pathogen detection methods, typically PCR, have numerous limitations, such as highly sophisticated equipment requirements, reagents, and trained personnel relying on well-established laboratories, besides being time-consuming. Thus, there is a crucial need to develop novel nucleic acid detection tools that are rapid, specific, sensitive, and cost-effective, particularly ones that can be used for versatile point-of-care diagnostic applications. Two new methods exploit unpredicted in vitro properties of CRISPR-Cas effectors, turning activated nucleases into basic amplifiers of a specific nucleic acid binding event. These effectors can be attached to a diversity of reporters and utilized in tandem with isothermal amplification approaches to create sensitive identification in multiple deployable field formats. Although still in their beginning, SHERLOCK and DETECTR technologies are potential methods for rapid detection and identification of infectious diseases, with ultrasensitive tests that do not require complicated processing. This review describes SHERLOCK and DETECTR technologies and assesses their properties, functions, and prospective to become the ultimate diagnostic tools for diagnosing infectious diseases and curbing disease outbreaks.
Keywords: CRISPR-Cas diagnostic tools; DETECTR; SHERLOCK; infectious diseases.
Copyright © 2021 Mustafa and Makhawi.
Figures
References
-
- Luthy IA, Ritacco V, Kantor IN. 2018. One hundred years after the “Spanish” flu (in Spanish). Medicina (B Aires) 78:113–118. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

