Identification of an Antiretroviral Small Molecule That Appears To Be a Host-Targeting Inhibitor of HIV-1 Assembly
- PMID: 33148797
- PMCID: PMC7925099
- DOI: 10.1128/JVI.00883-20
Identification of an Antiretroviral Small Molecule That Appears To Be a Host-Targeting Inhibitor of HIV-1 Assembly
Abstract
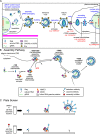
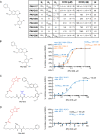
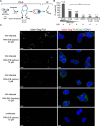
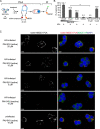
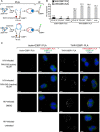
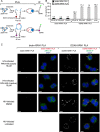
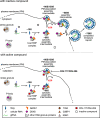
Given the projected increase in multidrug-resistant HIV-1, there is an urgent need for development of antiretrovirals that act on virus life cycle stages not targeted by drugs currently in use. Host-targeting compounds are of particular interest because they can offer a high barrier to resistance. Here, we report identification of two related small molecules that inhibit HIV-1 late events, a part of the HIV-1 life cycle for which potent and specific inhibitors are lacking. This chemotype was discovered using cell-free protein synthesis and assembly systems that recapitulate intracellular host-catalyzed viral capsid assembly pathways. These compounds inhibit replication of HIV-1 in human T cell lines and peripheral blood mononuclear cells, and are effective against a primary isolate. They reduce virus production, likely by inhibiting a posttranslational step in HIV-1 Gag assembly. Notably, the compound colocalizes with HIV-1 Gag in situ; however, unexpectedly, selection experiments failed to identify compound-specific resistance mutations in gag or pol, even though known resistance mutations developed upon parallel nelfinavir selection. Thus, we hypothesized that instead of binding to Gag directly, these compounds localize to assembly intermediates, the intracellular multiprotein complexes containing Gag and host factors that form during immature HIV-1 capsid assembly. Indeed, imaging of infected cells shows compound colocalized with two host enzymes found in assembly intermediates, ABCE1 and DDX6, but not two host proteins found in other complexes. While the exact target and mechanism of action of this chemotype remain to be determined, our findings suggest that these compounds represent first-in-class, host-targeting inhibitors of intracellular events in HIV-1 assembly.IMPORTANCE The success of antiretroviral treatment for HIV-1 is at risk of being undermined by the growing problem of drug resistance. Thus, there is a need to identify antiretrovirals that act on viral life cycle stages not targeted by drugs in use, such as the events of HIV-1 Gag assembly. To address this gap, we developed a compound screen that recapitulates the intracellular events of HIV-1 assembly, including virus-host interactions that promote assembly. This effort led to the identification of a new chemotype that inhibits HIV-1 replication at nanomolar concentrations, likely by acting on assembly. This compound colocalized with Gag and two host enzymes that facilitate capsid assembly. However, resistance selection did not result in compound-specific mutations in gag, suggesting that the chemotype does not directly target Gag. We hypothesize that this chemotype represents a first-in-class inhibitor of virus production that acts by targeting a virus-host complex important for HIV-1 Gag assembly.
Keywords: ABCE1; DDX6; HIV-1 assembly; HIV-1 capsid; RNA granules; antiretroviral agent; cell-free system; drug screen; virus-host interactions.
Copyright © 2021 Reed et al.
Figures








Similar articles
-
Addressing Antiretroviral Drug Resistance with Host-Targeting Drugs-First Steps towards Developing a Host-Targeting HIV-1 Assembly Inhibitor.Viruses. 2021 Mar 10;13(3):451. doi: 10.3390/v13030451. Viruses. 2021. PMID: 33802145 Free PMC article. Review.
-
Formation of RNA Granule-Derived Capsid Assembly Intermediates Appears To Be Conserved between Human Immunodeficiency Virus Type 1 and the Nonprimate Lentivirus Feline Immunodeficiency Virus.J Virol. 2018 Apr 13;92(9):e01761-17. doi: 10.1128/JVI.01761-17. Print 2018 May 1. J Virol. 2018. PMID: 29467316 Free PMC article.
-
Identifying the assembly intermediate in which Gag first associates with unspliced HIV-1 RNA suggests a novel model for HIV-1 RNA packaging.PLoS Pathog. 2018 Apr 17;14(4):e1006977. doi: 10.1371/journal.ppat.1006977. eCollection 2018 Apr. PLoS Pathog. 2018. PMID: 29664940 Free PMC article.
-
A temporospatial map that defines specific steps at which critical surfaces in the Gag MA and CA domains act during immature HIV-1 capsid assembly in cells.J Virol. 2014 May;88(10):5718-41. doi: 10.1128/JVI.03609-13. Epub 2014 Mar 12. J Virol. 2014. PMID: 24623418 Free PMC article.
-
HIV-1 Gag as an Antiviral Target: Development of Assembly and Maturation Inhibitors.Curr Top Med Chem. 2016;16(10):1154-66. doi: 10.2174/1568026615666150902102143. Curr Top Med Chem. 2016. PMID: 26329615 Free PMC article. Review.
Cited by
-
A pan-respiratory antiviral chemotype targeting a transient host multi-protein complex.Open Biol. 2024 Jun;14(6):230363. doi: 10.1098/rsob.230363. Epub 2024 Jun 19. Open Biol. 2024. PMID: 38889796 Free PMC article.
-
Addressing Antiretroviral Drug Resistance with Host-Targeting Drugs-First Steps towards Developing a Host-Targeting HIV-1 Assembly Inhibitor.Viruses. 2021 Mar 10;13(3):451. doi: 10.3390/v13030451. Viruses. 2021. PMID: 33802145 Free PMC article. Review.
-
Targeting the HIV-1 and HBV Capsids, an EnCore.Viruses. 2023 Mar 31;15(4):896. doi: 10.3390/v15040896. Viruses. 2023. PMID: 37112877 Free PMC article.
-
Developments in Exploring Fungal Secondary Metabolites as Antiviral Compounds and Advances in HIV-1 Inhibitor Screening Assays.Viruses. 2023 Apr 23;15(5):1039. doi: 10.3390/v15051039. Viruses. 2023. PMID: 37243125 Free PMC article. Review.
-
Small molecule protein assembly modulators with pan-cancer therapeutic efficacy.Open Biol. 2024 Dec;14(12):240210. doi: 10.1098/rsob.240210. Epub 2024 Dec 18. Open Biol. 2024. PMID: 39689856 Free PMC article.
References
-
- UNAIDS . 2019. Global HIV & AIDS statistics—2019 fact sheet. https://www.unaids.org/en/resources/fact-sheet.
-
- Boender TS, Sigaloff KC, McMahon JH, Kiertiburanakul S, Jordan MR, Barcarolo J, Ford N, Rinke de Wit TF, Bertagnolio S. 2015. Long-term virological outcomes of first-line antiretroviral therapy for HIV-1 in low- and middle-income countries: a systematic review and meta-analysis. Clin Infect Dis 61:1453–1461. doi:10.1093/cid/civ556. - DOI - PMC - PubMed
-
- Gregson J, Kaleebu P, Marconi VC, van Vuuren C, Ndembi N, Hamers RL, Kanki P, Hoffmann CJ, Lockman S, Pillay D, de Oliveira T, Clumeck N, Hunt G, Kerschberger B, Shafer RW, Yang C, Raizes E, Kantor R, Gupta RK. 2017. Occult HIV-1 drug resistance to thymidine analogues following failure of first-line tenofovir combined with a cytosine analogue and nevirapine or efavirenz in sub Saharan Africa: a retrospective multi-centre cohort study. Lancet Infect Dis 17:296–304. doi:10.1016/S1473-3099(16)30469-8. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

