Sediment Metagenomes as Time Capsules of Lake Microbiomes
- PMID: 33148818
- PMCID: PMC7643826
- DOI: 10.1128/mSphere.00512-20
Sediment Metagenomes as Time Capsules of Lake Microbiomes
Abstract
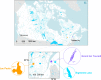
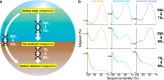
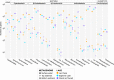
The reconstruction of ecological time series from lake sediment archives can retrace the environmental impact of human activities. Molecular genetic approaches in paleolimnology have provided unprecedented access to DNA time series, which record evidence of the microbial ecologies that underlaid historical lake ecosystems. Such studies often rely on single-gene surveys, and consequently, the full diversity of preserved microorganisms remains unexplored. In this study, we probed the diversity archived in contemporary and preindustrial sediments by comparative shotgun metagenomic analysis of surface water and sediment samples from three eastern Canadian lakes. In a strategy that was aimed at disentangling historical DNA from the indigenous sediment background, microbial preservation signals were captured by mapping sequence similarities between sediment metagenome reads and reference surface water metagenome assemblies. We detected preserved Cyanobacteria, diverse bacterioplankton, microeukaryotes, and viruses in sediment metagenomes. Among the preserved microorganisms were important groups never before reported in paleolimnological reconstructions, including bacteriophages (Caudovirales) and ubiquitous freshwater Betaproteobacteria (Polynucleobacter and Limnohabitans). In contrast, ultramicroscopic Actinobacteria ("Candidatus Nanopelagicales") and Alphaproteobacteria (Pelagibacterales) were apparently not well preserved in sediment metagenomes even though they were numerically dominant in surface water metagenomes. Overall, our study explored a novel application of whole-metagenome shotgun sequencing for discovering the DNA remains of a broad diversity of microorganisms preserved in lake sediments. The recovery of diverse microbial time series supports the taxonomic expansion of microbiome reconstructions and the development of novel microbial paleoindicators.IMPORTANCE Lakes are critical freshwater resources under mounting pressure from climate change and other anthropogenic stressors. The reconstruction of ecological time series from sediment archives with paleolimnological techniques has been shown to be an effective means of understanding how humans are modifying lake ecosystems over extended timescales. In this study, we combined shotgun DNA sequencing with a novel comparative analysis of surface water and sediment metagenomes to expose the diversity of microorganisms preserved in lake sediments. The detection of DNA from a broad diversity of preserved microbes serves to more fully reconstruct historical microbiomes and describe preimpact lake conditions.
Keywords: DNA preservation; bacterioplankton; metagenomics; paleogenomics; paleolimnology; shotgun sequencing; viruses.
Copyright © 2020 Garner et al.
Figures


Similar articles
-
A metagenomics roadmap to the uncultured genome diversity in hypersaline soda lake sediments.Microbiome. 2018 Sep 19;6(1):168. doi: 10.1186/s40168-018-0548-7. Microbiome. 2018. PMID: 30231921 Free PMC article.
-
More than 2500 years of oil exposure shape sediment microbiomes with the potential for syntrophic degradation of hydrocarbons linked to methanogenesis.Microbiome. 2017 Sep 11;5(1):118. doi: 10.1186/s40168-017-0337-8. Microbiome. 2017. PMID: 28893308 Free PMC article.
-
Distinctive Patterns in the Taxonomical Resolution of Bacterioplankton in the Sediment and Pore Waters of Contrasted Freshwater Lakes.Microb Ecol. 2018 Apr;75(3):662-673. doi: 10.1007/s00248-017-1074-z. Epub 2017 Sep 17. Microb Ecol. 2018. PMID: 28920165
-
Environmental paleomicrobiology: using DNA preserved in aquatic sediments to its full potential.Environ Microbiol. 2022 May;24(5):2201-2209. doi: 10.1111/1462-2920.15913. Epub 2022 Feb 7. Environ Microbiol. 2022. PMID: 35049133 Free PMC article. Review.
-
Ecological and functional roles of bacteriophages in contrasting environments: marine, terrestrial and human gut.Curr Opin Microbiol. 2022 Dec;70:102229. doi: 10.1016/j.mib.2022.102229. Epub 2022 Nov 5. Curr Opin Microbiol. 2022. PMID: 36347213 Review.
Cited by
-
Red mark syndrome: Is the aquaculture water microbiome a keystone for understanding the disease aetiology?Front Microbiol. 2023 Feb 27;14:1059127. doi: 10.3389/fmicb.2023.1059127. eCollection 2023. Front Microbiol. 2023. PMID: 36922974 Free PMC article.
-
Lake microbiome and trophy fluctuations of the ancient hemp rettery.Sci Rep. 2022 May 25;12(1):8846. doi: 10.1038/s41598-022-12761-w. Sci Rep. 2022. PMID: 35614182 Free PMC article.
-
Landscape Setting Drives the Microbial Eukaryotic Community Structure in Four Swedish Mountain Lakes over the Holocene.Microorganisms. 2021 Feb 11;9(2):355. doi: 10.3390/microorganisms9020355. Microorganisms. 2021. PMID: 33670228 Free PMC article.
-
Postglacial bioweathering, soil nutrient cycling, and podzolization from palaeometagenomics of plants, fungi, and bacteria.Sci Adv. 2025 May 9;11(19):eadj5527. doi: 10.1126/sciadv.adj5527. Epub 2025 May 7. Sci Adv. 2025. PMID: 40333974 Free PMC article.
-
Large scale exploration reveals rare taxa crucially shape microbial assembly in alkaline lake sediments.NPJ Biofilms Microbiomes. 2024 Jul 28;10(1):62. doi: 10.1038/s41522-024-00537-1. NPJ Biofilms Microbiomes. 2024. PMID: 39069527 Free PMC article.
References
-
- Tranvik LJ, Downing JA, Cotner JB, Loiselle SA, Striegl RG, Ballatore TJ, Dillon P, Finlay K, Fortino K, Knoll LB, Kortelainen PL, Kutser T, Larsen S, Laurion I, Leech DM, Leigh McCallister S, McKnight DM, Melack JM, Overholt E, Porter JA, Prairie Y, Renwick WH, Roland F, Sherman BS, Schindler DW, Sobek S, Tremblay A, Vanni MJ, Verschoor AM, Von Wachenfeldt E, Weyhenmeyer GA. 2009. Lakes and reservoirs as regulators of carbon cycling and climate. Limnol Oceanogr 54:2298–2314. doi:10.4319/lo.2009.54.6_part_2.2298. - DOI
-
- Williamson CE, Dodds W, Kratz TK, Palmer MA. 2008. Lakes and streams as sentinels of environmental change in terrestrial and atmospheric processes. Front Ecol Environ 6:247–254. doi:10.1890/070140. - DOI
-
- Reid AJ, Carlson AK, Creed IF, Eliason EJ, Gell PA, Johnson PTJ, Kidd KA, MacCormack TJ, Olden JD, Ormerod SJ, Smol JP, Taylor WW, Tockner K, Vermaire JC, Dudgeon D, Cooke SJ. 2019. Emerging threats and persistent conservation challenges for freshwater biodiversity. Biol Rev Camb Philos Soc 94:849–873. doi:10.1111/brv.12480. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
