Spo0A Suppresses sin Locus Expression in Clostridioides difficile
- PMID: 33148827
- PMCID: PMC7643835
- DOI: 10.1128/mSphere.00963-20
Spo0A Suppresses sin Locus Expression in Clostridioides difficile
Abstract
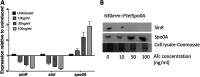
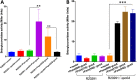
Clostridioides difficile is the leading cause of nosocomial infection and is the causative agent of antibiotic-associated diarrhea. The severity of the disease is directly associated with toxin production, and spores are responsible for the transmission and persistence of the organism. Previously, we characterized sin locus regulators SinR and SinR' (we renamed it SinI), where SinR is the regulator of toxin production and sporulation. The SinI regulator acts as its antagonist. In Bacillus subtilis, Spo0A, the master regulator of sporulation, controls SinR by regulating the expression of its antagonist, sinI However, the role of Spo0A in the expression of sinR and sinI in C. difficile had not yet been reported. In this study, we tested spo0A mutants in three different C. difficile strains, R20291, UK1, and JIR8094, to understand the role of Spo0A in sin locus expression. Western blot analysis revealed that spo0A mutants had increased SinR levels. Quantitative reverse transcription-PCR (qRT-PCR) analysis of its expression further supported these data. By carrying out genetic and biochemical assays, we show that Spo0A can bind to the upstream region of this locus to regulates its expression. This study provides vital information that Spo0A regulates the sin locus, which controls critical pathogenic traits such as sporulation, toxin production, and motility in C. difficileIMPORTANCEClostridioides difficile is the leading cause of antibiotic-associated diarrheal disease in the United States. During infection, C. difficile spores germinate, and the vegetative bacterial cells produce toxins that damage host tissue. In C. difficile, the sin locus is known to regulate both sporulation and toxin production. In this study, we show that Spo0A, the master regulator of sporulation, controls sin locus expression. Results from our study suggest that Spo0A directly regulates the expression of this locus by binding to its upstream DNA region. This observation adds new detail to the gene regulatory network that connects sporulation and toxin production in this pathogen.
Keywords: C. difficile; Clostridioides difficile; SinR; Spo0A; gene regulation; virulence gene regulation.
Copyright © 2020 Dhungel and Govind.
Figures


Similar articles
-
Pleiotropic roles of Clostridium difficile sin locus.PLoS Pathog. 2018 Mar 12;14(3):e1006940. doi: 10.1371/journal.ppat.1006940. eCollection 2018 Mar. PLoS Pathog. 2018. PMID: 29529083 Free PMC article.
-
Clostridioides difficile SinR' regulates toxin, sporulation and motility through protein-protein interaction with SinR.Anaerobe. 2019 Oct;59:1-7. doi: 10.1016/j.anaerobe.2019.05.002. Epub 2019 May 8. Anaerobe. 2019. PMID: 31077800 Free PMC article.
-
C. difficile 630Δerm Spo0A regulates sporulation, but does not contribute to toxin production, by direct high-affinity binding to target DNA.PLoS One. 2012;7(10):e48608. doi: 10.1371/journal.pone.0048608. Epub 2012 Oct 31. PLoS One. 2012. PMID: 23119071 Free PMC article.
-
Regulatory networks: Linking toxin production and sporulation in Clostridioides difficile.Anaerobe. 2025 Feb;91:102920. doi: 10.1016/j.anaerobe.2024.102920. Epub 2024 Nov 7. Anaerobe. 2025. PMID: 39521117 Review.
-
An evolutionary link between sporulation and prophage induction in the structure of a repressor:anti-repressor complex.J Mol Biol. 1998 Nov 13;283(5):907-12. doi: 10.1006/jmbi.1998.2163. J Mol Biol. 1998. PMID: 9799632 Review.
Cited by
-
Regulation of Clostridial Toxin Gene Expression: A Pasteurian Tradition.Toxins (Basel). 2023 Jun 26;15(7):413. doi: 10.3390/toxins15070413. Toxins (Basel). 2023. PMID: 37505682 Free PMC article. Review.
-
Regulation of Clostridioides difficile toxin production.Curr Opin Microbiol. 2022 Feb;65:95-100. doi: 10.1016/j.mib.2021.10.018. Epub 2021 Nov 12. Curr Opin Microbiol. 2022. PMID: 34781095 Free PMC article. Review.
-
Regulatory Role of Anti-Sigma Factor RsbW in Clostridioides difficile Stress Response, Persistence, and Infection.J Bacteriol. 2023 May 25;205(5):e0046622. doi: 10.1128/jb.00466-22. Epub 2023 Apr 26. J Bacteriol. 2023. PMID: 37098979 Free PMC article.
-
A conserved switch controls virulence, sporulation, and motility in C. difficile.PLoS Pathog. 2024 May 13;20(5):e1012224. doi: 10.1371/journal.ppat.1012224. eCollection 2024 May. PLoS Pathog. 2024. PMID: 38739653 Free PMC article.
-
Fight or flee, a vital choice for Clostridioides difficile.mLife. 2024 Feb 9;3(1):14-20. doi: 10.1002/mlf2.12102. eCollection 2024 Mar. mLife. 2024. PMID: 38827507 Free PMC article.
References
-
- Prete RD, Ronga L, Addati G, Magrone R, Abbasciano A, Decimo M, Miragliotta G. 2019. Clostridium difficile. a review on an emerging infection. La Clinica Terapeutica 1:e41–e47. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
