NaCl Concentration-Dependent Aminoglycoside Resistance of Halomonas socia CKY01 and Identification of Related Genes
- PMID: 33148940
- PMCID: PMC9705875
- DOI: 10.4014/jmb.2009.09017
NaCl Concentration-Dependent Aminoglycoside Resistance of Halomonas socia CKY01 and Identification of Related Genes
Abstract
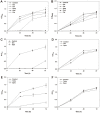
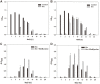
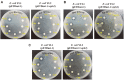
Among various species of marine bacteria, those belonging to the genus Halomonas have several promising applications and have been studied well. However, not much information has been available on their antibiotic resistance. In our efforts to learn about the antibiotic resistance of strain Halomonas socia CKY01, which showed production of various hydrolases and growth promotion by osmolytes in previous study, we found that it exhibited resistance to multiple antibiotics including kanamycin, ampicillin, oxacillin, carbenicillin, gentamicin, apramycin, tetracycline, and spectinomycin. However, the H. socia CKY01 resistance pattern to kanamycin, gentamicin, apramycin, tetracycline, and spectinomycin differed in the presence of 10% NaCl and 1% NaCl in the culture medium. To determine the mechanism underlying this NaCl concentration-dependent antibiotic resistance, we compared four aminoglycoside resistance genes under different salt conditions while also performing time-dependent reverse transcription PCR. We found that the aph2 gene encoding aminoglycoside phosphotransferase showed increased expression under the 10% rather than 1% NaCl conditions. When these genes were overexpressed in an Escherichia coli strain, pETDuet-1::aph2 showed a smaller inhibition zone in the presence of kanamycin, gentamicin, and apramycin than the respective control, suggesting aph2 was involved in aminoglycoside resistance. Our results demonstrated a more direct link between NaCl and aminoglycoside resistance exhibited by the H. socia CKY01 strain.
Keywords: Halomonas strain; antibiotic resistance; genome sequencing; salt tolerance.
Conflict of interest statement
The authors have no financial conflicts of interest to declare.
Figures

References
-
- Gurav R, Bhatia SK, Moon Y-M, Choi T-R, Jung H-R, Yang S-Y, et al. One-pot exploitation of chitin biomass for simultaneous production of electricity, n-acetylglucosamine and polyhydroxyalkanoates in microbial fuel cell using novel marine bacterium Arenibacter palladensis YHY2. J. Clean. Prod. 2019;209:324–332. doi: 10.1016/j.jclepro.2018.10.252. - DOI
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

