Polyandry blocks gene drive in a wild house mouse population
- PMID: 33149121
- PMCID: PMC7643059
- DOI: 10.1038/s41467-020-18967-8
Polyandry blocks gene drive in a wild house mouse population
Abstract
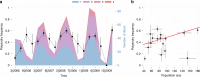
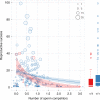
Gene drives are genetic elements that manipulate Mendelian inheritance ratios in their favour. Understanding the forces that explain drive frequency in natural populations is a long-standing focus of evolutionary research. Recently, the possibility to create artificial drive constructs to modify pest populations has exacerbated our need to understand how drive spreads in natural populations. Here, we study the impact of polyandry on a well-known gene drive, called t haplotype, in an intensively monitored population of wild house mice. First, we show that house mice are highly polyandrous: 47% of 682 litters were sired by more than one male. Second, we find that drive-carrying males are particularly compromised in sperm competition, resulting in reduced reproductive success. As a result, drive frequency decreased during the 4.5 year observation period. Overall, we provide the first direct evidence that the spread of a gene drive is hampered by reproductive behaviour in a natural population.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Burt, A. & Trivers, R. Genes in Conflict: The Biology of Selfish Genetic Elements (Belknap Press, Cambridge, 2006).
-
- Lindholm AK, et al. The ecology and evolutionary dynamics of meiotic drive. Trends Ecol. Evolution. 2016;31:316–326. - PubMed
-
- Champer J, Buchman A, Akbari OS. Cheating evolution: engineering gene drives to manipulate the fate of wild populations. Nat. Rev. Genet. 2016;17:146. - PubMed
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Research Materials

