Type I IFN exacerbates disease in tuberculosis-susceptible mice by inducing neutrophil-mediated lung inflammation and NETosis
- PMID: 33149141
- PMCID: PMC7643080
- DOI: 10.1038/s41467-020-19412-6
Type I IFN exacerbates disease in tuberculosis-susceptible mice by inducing neutrophil-mediated lung inflammation and NETosis
Abstract
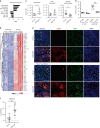
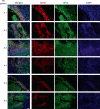
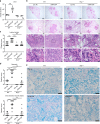
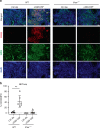
Tuberculosis (TB) is a leading cause of mortality due to infectious disease, but the factors determining disease progression are unclear. Transcriptional signatures associated with type I IFN signalling and neutrophilic inflammation were shown to correlate with disease severity in mouse models of TB. Here we show that similar transcriptional signatures correlate with increased bacterial loads and exacerbate pathology during Mycobacterium tuberculosis infection upon GM-CSF blockade. Loss of GM-CSF signalling or genetic susceptibility to TB (C3HeB/FeJ mice) result in type I IFN-induced neutrophil extracellular trap (NET) formation that promotes bacterial growth and promotes disease severity. Consistently, NETs are present in necrotic lung lesions of TB patients responding poorly to antibiotic therapy, supporting the role of NETs in a late stage of TB pathogenesis. Our findings reveal an important cytokine-based innate immune effector network with a central role in determining the outcome of M. tuberculosis infection.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- WHO. Global Tuberculosis Report 2019. (World Health Organization, 2019).
Publication types
MeSH terms
Substances
Grants and funding
- FC001129/MRC_/Medical Research Council/United Kingdom
- FC001999/CRUK_/Cancer Research UK/United Kingdom
- MC_U117565642/MRC_/Medical Research Council/United Kingdom
- U117565642/MRC_/Medical Research Council/United Kingdom
- FC001999/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- FC001126/WT_/Wellcome Trust/United Kingdom
- FC001129/WT_/Wellcome Trust/United Kingdom
- FC001126/MRC_/Medical Research Council/United Kingdom
- FC001129/CRUK_/Cancer Research UK/United Kingdom
- FC001999/WT_/Wellcome Trust/United Kingdom
- FC001126/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

