Subgroup level differences of physiological activities in marine Lokiarchaeota
- PMID: 33149207
- PMCID: PMC8027215
- DOI: 10.1038/s41396-020-00818-5
Subgroup level differences of physiological activities in marine Lokiarchaeota
Abstract
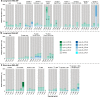
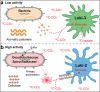
Asgard is a recently discovered archaeal superphylum, closely linked to the emergence of eukaryotes. Among Asgard archaea, Lokiarchaeota are abundant in marine sediments, but their in situ activities are largely unknown except for Candidatus 'Prometheoarchaeum syntrophicum'. Here, we tracked the activity of Lokiarchaeota in incubations with Helgoland mud area sediments (North Sea) by stable isotope probing (SIP) with organic polymers, 13C-labelled inorganic carbon, fermentation intermediates and proteins. Within the active archaea, we detected members of the Lokiarchaeota class Loki-3, which appeared to mixotrophically participate in the degradation of lignin and humic acids while assimilating CO2, or heterotrophically used lactate. In contrast, members of the Lokiarchaeota class Loki-2 utilized protein and inorganic carbon, and degraded bacterial biomass formed in incubations. Metagenomic analysis revealed pathways for lactate degradation, and involvement in aromatic compound degradation in Loki-3, while the less globally distributed Loki-2 instead rely on protein degradation. We conclude that Lokiarchaeotal subgroups vary in their metabolic capabilities despite overlaps in their genomic equipment, and suggest that these subgroups occupy different ecologic niches in marine sediments.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
-
- Zaremba-Niedzwiedzka K, Caceres EF, Saw JH, Backstrom D, Juzokaite L, Vancaester E, et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature. 2017;541:353–8. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

