Characterisation of the main PSA glycoforms in aggressive prostate cancer
- PMID: 33149259
- PMCID: PMC7643140
- DOI: 10.1038/s41598-020-75526-3
Characterisation of the main PSA glycoforms in aggressive prostate cancer
Abstract
Serum levels of prostate specific antigen (PSA) are commonly used for prostate cancer (PCa) detection. However, their lack of specificity to distinguish benign prostate pathologies from PCa, or indolent from aggressive PCa have prompted the study of new non-invasive PCa biomarkers. Aberrant glycosylation is involved in neoplastic progression and specific changes in PSA glycosylation pattern, as the reduction in the percentage of α2,6-sialic acid (SA) are associated with PCa aggressiveness. In this study, we have characterised the main sialylated PSA glycoforms from blood serum of aggressive PCa patients and have compared with those of standard PSA from healthy individuals' seminal plasma. PSA was immunoprecipitated and α2,6-SA were separated from α2,3-SA glycoforms using SNA affinity chromatography. PSA N-glycans were released, labelled and analysed by hydrophilic interaction liquid chromatography combined with exoglycosidase digestions. The results showed that blood serum PSA sialylated glycoforms containing GalNAc residues were largely increased in aggressive PCa patients, whereas the disialylated core fucosylated biantennary structures with α2,6-SA, which are the major PSA glycoforms in standard PSA from healthy individuals, were markedly reduced in aggressive PCa. The identification of these main PSA glycoforms altered in aggressive PCa opens the way to design specific strategies to target them, which will be useful to improve PCa risk stratification.
Conflict of interest statement
The authors declare no competing interests.
Figures
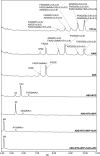
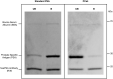
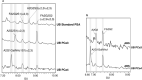

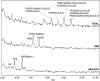
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

