Neuronal diversity and convergence in a visual system developmental atlas
- PMID: 33149298
- PMCID: PMC7790857
- DOI: 10.1038/s41586-020-2879-3
Neuronal diversity and convergence in a visual system developmental atlas
Abstract
Deciphering how neuronal diversity is established and maintained requires a detailed knowledge of neuronal gene expression throughout development. In contrast to mammalian brains1,2, the large neuronal diversity of the Drosophila optic lobe3 and its connectome4-6 are almost completely characterized. However, a molecular characterization of this neuronal diversity, particularly during development, has been lacking. Here we present insights into brain development through a nearly complete description of the transcriptomic diversity of the optic lobes of Drosophila. We acquired the transcriptome of 275,000 single cells at adult and at five pupal stages, and built a machine-learning framework to assign them to almost 200 cell types at all time points during development. We discovered two large neuronal populations that wrap neuropils during development but die just before adulthood, as well as neuronal subtypes that partition dorsal and ventral visual circuits by differential Wnt signalling throughout development. Moreover, we show that the transcriptomes of neurons that are of the same type but are produced days apart become synchronized shortly after their production. During synaptogenesis we also resolved neuronal subtypes that, although differing greatly in morphology and connectivity, converge to indistinguishable transcriptomic profiles in adults. Our datasets almost completely account for the known neuronal diversity of the Drosophila optic lobes, and serve as a paradigm to understand brain development across species.
Conflict of interest statement
Declaration of Interests
Authors declare no conflicts of interest.
Figures



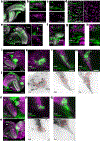






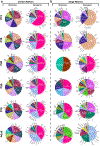
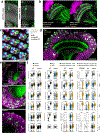





References
-
- Fischbach KF & Dittrich APM The optic lobe of Drosophila melanogaster. I. A Golgi analysis of wild-type structure. Cell Tissue Res. 258, 441–475, doi:10.1007/BF00218858 (1989). - DOI
References (online-only)
-
- Srivastava N, Hinton G, Krizhevsky A, Sutskever I & Salakhutdinov R Dropout: A Simple Way to Prevent Neural Networks from Overfitting. Vol. 15 (2014).
-
- Yarin G & Zoubin G 1050–1059 (PMLR, 2016).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

