Inhibition of LTβR signalling activates WNT-induced regeneration in lung
- PMID: 33149305
- PMCID: PMC7718297
- DOI: 10.1038/s41586-020-2882-8
Inhibition of LTβR signalling activates WNT-induced regeneration in lung
Erratum in
-
Publisher Correction: Inhibition of LTβR signalling activates WNT-induced regeneration in lung.Nature. 2021 Jan;589(7842):E6. doi: 10.1038/s41586-020-03087-6. Nature. 2021. PMID: 33353984 No abstract available.
Abstract
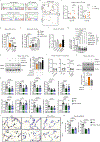
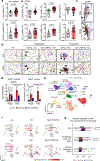
Lymphotoxin β-receptor (LTβR) signalling promotes lymphoid neogenesis and the development of tertiary lymphoid structures1,2, which are associated with severe chronic inflammatory diseases that span several organ systems3-6. How LTβR signalling drives chronic tissue damage particularly in the lung, the mechanism(s) that regulate this process, and whether LTβR blockade might be of therapeutic value have remained unclear. Here we demonstrate increased expression of LTβR ligands in adaptive and innate immune cells, enhanced non-canonical NF-κB signalling, and enriched LTβR target gene expression in lung epithelial cells from patients with smoking-associated chronic obstructive pulmonary disease (COPD) and from mice chronically exposed to cigarette smoke. Therapeutic inhibition of LTβR signalling in young and aged mice disrupted smoking-related inducible bronchus-associated lymphoid tissue, induced regeneration of lung tissue, and reverted airway fibrosis and systemic muscle wasting. Mechanistically, blockade of LTβR signalling dampened epithelial non-canonical activation of NF-κB, reduced TGFβ signalling in airways, and induced regeneration by preventing epithelial cell death and activating WNT/β-catenin signalling in alveolar epithelial progenitor cells. These findings suggest that inhibition of LTβR signalling represents a viable therapeutic option that combines prevention of tertiary lymphoid structures1 and inhibition of apoptosis with tissue-regenerative strategies.
Conflict of interest statement
All other authors declare no competing interests.
Data Availability
Microarray data was submitted to the NCBI Gene Expression Omnibus (GEO) database (
Code Availability
All code used for data visualization of the single cell RNA-Seq data can be found at
Figures











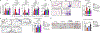

References
Method’s References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

