KMT2B-related disorders: expansion of the phenotypic spectrum and long-term efficacy of deep brain stimulation
- PMID: 33150406
- PMCID: PMC7719027
- DOI: 10.1093/brain/awaa304
KMT2B-related disorders: expansion of the phenotypic spectrum and long-term efficacy of deep brain stimulation
Abstract
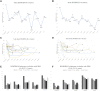
Heterozygous mutations in KMT2B are associated with an early-onset, progressive and often complex dystonia (DYT28). Key characteristics of typical disease include focal motor features at disease presentation, evolving through a caudocranial pattern into generalized dystonia, with prominent oromandibular, laryngeal and cervical involvement. Although KMT2B-related disease is emerging as one of the most common causes of early-onset genetic dystonia, much remains to be understood about the full spectrum of the disease. We describe a cohort of 53 patients with KMT2B mutations, with detailed delineation of their clinical phenotype and molecular genetic features. We report new disease presentations, including atypical patterns of dystonia evolution and a subgroup of patients with a non-dystonic neurodevelopmental phenotype. In addition to the previously reported systemic features, our study has identified co-morbidities, including the risk of status dystonicus, intrauterine growth retardation, and endocrinopathies. Analysis of this study cohort (n = 53) in tandem with published cases (n = 80) revealed that patients with chromosomal deletions and protein truncating variants had a significantly higher burden of systemic disease (with earlier onset of dystonia) than those with missense variants. Eighteen individuals had detailed longitudinal data available after insertion of deep brain stimulation for medically refractory dystonia. Median age at deep brain stimulation was 11.5 years (range: 4.5-37.0 years). Follow-up after deep brain stimulation ranged from 0.25 to 22 years. Significant improvement of motor function and disability (as assessed by the Burke Fahn Marsden's Dystonia Rating Scales, BFMDRS-M and BFMDRS-D) was evident at 6 months, 1 year and last follow-up (motor, P = 0.001, P = 0.004, and P = 0.012; disability, P = 0.009, P = 0.002 and P = 0.012). At 1 year post-deep brain stimulation, >50% of subjects showed BFMDRS-M and BFMDRS-D improvements of >30%. In the long-term deep brain stimulation cohort (deep brain stimulation inserted for >5 years, n = 8), improvement of >30% was maintained in 5/8 and 3/8 subjects for the BFMDRS-M and BFMDRS-D, respectively. The greatest BFMDRS-M improvements were observed for trunk (53.2%) and cervical (50.5%) dystonia, with less clinical impact on laryngeal dystonia. Improvements in gait dystonia decreased from 20.9% at 1 year to 16.2% at last assessment; no patient maintained a fully independent gait. Reduction of BFMDRS-D was maintained for swallowing (52.9%). Five patients developed mild parkinsonism following deep brain stimulation. KMT2B-related disease comprises an expanding continuum from infancy to adulthood, with early evidence of genotype-phenotype correlations. Except for laryngeal dysphonia, deep brain stimulation provides a significant improvement in quality of life and function with sustained clinical benefit depending on symptoms distribution.
Keywords: KMT2B; deep brain stimulation (DBS); dystonia; genetics; neurodevelopment.
© The Author(s) (2020). Published by Oxford University Press on behalf of the Guarantors of Brain. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures


Update of
-
KMT2B-related disorders: expansion of the phenotypic spectrum and long-term efficacy of deep brain stimulation.ArXiv [Preprint]. 2025 Feb 10:arXiv:2502.06320v1. ArXiv. 2025. Update in: Brain. 2020 Dec 5;143(11):3242-3261. doi: 10.1093/brain/awaa304. PMID: 39990802 Free PMC article. Updated. Preprint.
References
-
- Baizabal-Carvallo JF, Alonso-Juarez M.. Generalized dystonia associated with mutation in the histone methyltransferase gene KMT2B (DYT28) and white matter abnormalities. Parkinsonism Relat Disord 2018; 49: 116–7. - PubMed
-
- Ben-Haim S, Flatow V, Cheung T, Cho C, Tagliati M, Alterman RL.. Deep brain stimulation for status dystonicus: a case series and review of the literature. Stereotact Funct Neurosurg 2016; 94: 207–15. - PubMed
-
- Bereket A, Turan S, Alper G, Comu S, Alpay H, Akalin F.. Two patients with Kabuki syndrome presenting with endocrine problems. J Pediatr Endocrinol Metab 2001; 14: 215–20. - PubMed
-
- Blanchard A, Ea V, Roubertie A, Martin M, Coquart C, Claustres M, et al. DYT6 dystonia: review of the literature and creation of the UMD Locus-Specific Database (LSDB) for mutations in the THAP1 gene. Hum Mutat 2011; 32: 1213–24. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

