Frontotemporal dementia-linked P112H mutation of TDP-43 induces protein structural change and impairs its RNA binding function
- PMID: 33151007
- PMCID: PMC7784771
- DOI: 10.1002/pro.3990
Frontotemporal dementia-linked P112H mutation of TDP-43 induces protein structural change and impairs its RNA binding function
Abstract
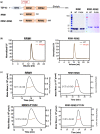
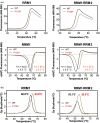
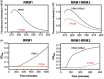
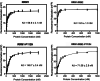
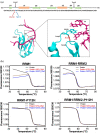
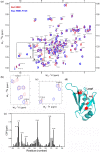
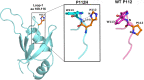
TDP-43 forms the primary constituents of the cytoplasmic inclusions contributing to various neurodegenerative diseases, including amyotrophic lateral sclerosis and frontotemporal dementia (FTD). Over 60 TDP-43 mutations have been identified in patients suffering from these two diseases, but most variations are located in the protein's disordered C-terminal glycine-rich region. P112H mutation of TDP-43 has been uniquely linked to FTD, and is located in the first RNA recognition motif (RRM1). This mutation is thought to be pathogenic, but its impact on TDP-43 at the protein level remains unclear. Here, we compare the biochemical and biophysical properties of TDP-43 truncated proteins with or without P112H mutation. We show that P112H-mutated TDP-43 proteins exhibit higher thermal stability, impaired RNA-binding activity, and a reduced tendency to aggregate relative to wild-type proteins. Near-UV CD, 2D-nuclear-magnetic resonance, and intrinsic fluorescence spectrometry further reveal that the P112H mutation in RRM1 generates local conformational changes surrounding the mutational site that disrupt the stacking interactions of the W113 side chain with nucleic acids. Together, these results support the notion that P112H mutation of TDP-43 contributes to FTD through functional impairment of RNA metabolism and/or structural changes that curtail protein clearance.
Keywords: RNA recognition motif; RNA-binding protein; neurodegenerative disease; protein aggregation.
© 2020 The Authors. Protein Science published by Wiley Periodicals LLC on behalf of The Protein Society.
Figures

References
-
- Vanden Broeck L, Callaerts P, Dermaut B. TDP‐43‐mediated neurodegeneration: Towards a loss‐of‐function hypothesis? Trends Mol Med. 2014;20:66–71. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

