Algorithmic assessment of missense mutation severity in the Von-Hippel Lindau protein
- PMID: 33151962
- PMCID: PMC7644048
- DOI: 10.1371/journal.pone.0234100
Algorithmic assessment of missense mutation severity in the Von-Hippel Lindau protein
Abstract
Von Hippel-Lindau disease (VHL) is an autosomal dominant rare disease that causes the formation of angiogenic tumors. When functional, pVHL acts as an E3 ubiquitin ligase that negatively regulates hypoxia inducible factor (HIF). Genetic mutations that perturb the structure of pVHL result in dysregulation of HIF, causing a wide array of tumor pathologies including retinal angioma, pheochromocytoma, central nervous system hemangioblastoma, and clear cell renal carcinoma. These VHL-related cancers occur throughout the lifetime of the patient, requiring frequent intervention procedures, such as surgery, to remove the tumors. Although VHL is classified as a rare disease (1 in 39,000 to 1 in 91,000 affected) there is a large heterogeneity in genetic mutations listed for observed pathologies. Understanding how these specific mutations correlate with the myriad of observed pathologies for VHL could provide clinicians insight into the potential severity and onset of disease. Using a select set of 285 ClinVar mutations in VHL, we developed a multiparametric scoring algorithm to evaluate the overall clinical severity of missense mutations in pVHL. The mutations were assessed according to eight weighted parameters as a comprehensive evaluation of protein misfolding and malfunction. Higher mutation scores were strongly associated with pathogenicity. Our approach establishes a novel in silico method by which VHL-specific mutations can be assessed for their severity and effect on the biophysical functions of the VHL protein.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

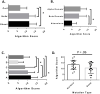
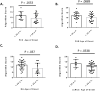
Similar articles
-
von Hippel-Lindau disease: a clinical and scientific review.Eur J Hum Genet. 2011 Jun;19(6):617-23. doi: 10.1038/ejhg.2010.175. Epub 2011 Mar 9. Eur J Hum Genet. 2011. PMID: 21386872 Free PMC article. Review.
-
Genotype and phenotype correlation in von Hippel-Lindau disease based on alteration of the HIF-α binding site in VHL protein.Genet Med. 2018 Oct;20(10):1266-1273. doi: 10.1038/gim.2017.261. Epub 2018 Mar 29. Genet Med. 2018. PMID: 29595810
-
Contrasting effects on HIF-1alpha regulation by disease-causing pVHL mutations correlate with patterns of tumourigenesis in von Hippel-Lindau disease.Hum Mol Genet. 2001 May 1;10(10):1029-38. doi: 10.1093/hmg/10.10.1029. Hum Mol Genet. 2001. PMID: 11331613
-
Comprehensive characterization of a Canadian cohort of von Hippel-Lindau disease patients.Clin Genet. 2019 Nov;96(5):461-467. doi: 10.1111/cge.13613. Epub 2019 Aug 6. Clin Genet. 2019. PMID: 31368132
-
The HIF and other quandaries in VHL disease.Oncogene. 2018 Jan 11;37(2):139-147. doi: 10.1038/onc.2017.338. Epub 2017 Sep 18. Oncogene. 2018. PMID: 28925400 Review.
Cited by
-
Germline Pathogenic Variants Identified by Targeted Next-Generation Sequencing of Susceptibility Genes in Pheochromocytoma and Paraganglioma.J Kidney Cancer VHL. 2021 Mar 13;8(1):19-24. doi: 10.15586/jkcvhl.v8i1.171. eCollection 2021. J Kidney Cancer VHL. 2021. PMID: 33777662 Free PMC article.
-
The germline mutational landscape of genitourinary cancers and its indication for prognosis and risk.BMC Urol. 2022 Nov 30;22(1):196. doi: 10.1186/s12894-022-01141-1. BMC Urol. 2022. PMID: 36451132 Free PMC article.
-
The genetic differences between types 1 and 2 in von Hippel-Lindau syndrome: comprehensive meta-analysis.BMC Ophthalmol. 2024 Aug 13;24(1):343. doi: 10.1186/s12886-024-03597-1. BMC Ophthalmol. 2024. PMID: 39138406 Free PMC article.
References
-
- Gossage L, Eisen T, Maher ER. VHL, the story of a tumour suppressor gene [Internet]. Vol. 15, Nature Reviews Cancer. 2015. [cited 2019 Sep 11]. p. 55–64. Available from: www.nature.com/reviews/cancer. 10.1038/nrc3844 - DOI - PubMed
-
- Haase V. The VHL Tumor Suppressor: Master Regulator of HIF. Curr Pharm Des [Internet]. 2009. [cited 2019 Oct 9];15(33):3895–903. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3622710/pdf/nihms454216.pdf. 10.2174/138161209789649394 - DOI - PMC - PubMed
-
- Tory K, Brauch H, Linehan M, Barba D, Oldfield E, Katz MF, et al. Specific genetic change in tumors associated with von hippel-lindau disease. J Natl Cancer Inst [Internet]. 1989. July 19 [cited 2019 Oct 10];81(14):1097–101. Available from: https://academic.oup.com/jnci/article-lookup/doi/10.1093/jnci/81.14.1097. - DOI - PubMed
-
- Crossey PA, Foster K, Richards FM, Phipps ME, Latif F, Tory K, et al. Molecular genetic investigations of the mechanism of tumourigenesis in von Hippel-Lindau disease: analysis of allele loss in VHL tumours. Hum Genet [Internet]. 1994. January [cited 2019 Oct 10];93(1):53–8. Available from: http://www.ncbi.nlm.nih.gov/pubmed/8270255. 10.1007/BF00218913 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

