PDIA3 Expression in Glioblastoma Modulates Macrophage/Microglia Pro-Tumor Activation
- PMID: 33153019
- PMCID: PMC7662700
- DOI: 10.3390/ijms21218214
PDIA3 Expression in Glioblastoma Modulates Macrophage/Microglia Pro-Tumor Activation
Abstract
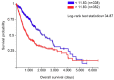
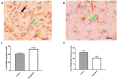
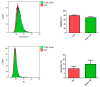
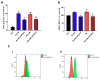
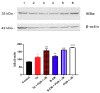
The glioblastoma (GB) microenvironment includes cells of the innate immune system identified as glioma-associated microglia/macrophages (GAMs) that are still poorly characterized. A potential role on the mechanisms regulating GAM activity might be played by the endoplasmic reticulum protein ERp57/PDIA3 (protein disulfide-isomerase A3), the modulation of which has been reported in a variety of cancers. Moreover, by using The Cancer Genome Atlas database, we found that overexpression of PDIA3 correlated with about 55% reduction of overall survival of glioma patients. Therefore, we analyzed the expression of ERp57/PDIA3 using specimens obtained after surgery from 18 GB patients. Immunohistochemical analysis of tumor samples revealed ERp57/PDIA3 expression in GB cells as well as in GAMs. The ERp57/PDIA3 levels were higher in GAMs than in the microglia present in the surrounding parenchyma. Therefore, we studied the role of PDIA3 modulation in microglia-glioma interaction, based on the ability of conditioned media collected from human GB cells to induce the activation of microglial cells. The results indicated that reduced PDIA3 expression/activity in GB cells significantly limited the microglia pro-tumor polarization towards the M2 phenotype and the production of pro-inflammatory factors. Our data support a role of PDIA3 expression in GB-mediated protumor activation of microglia.
Keywords: IL6; PDIA3; STAT3; glioma; microglia; punicalagin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







Similar articles
-
Molecular profiling of the tumor microenvironment in glioblastoma patients: correlation of microglia/macrophage polarization state with metalloprotease expression profiles and survival.Biosci Rep. 2019 Jun 20;39(6):BSR20182361. doi: 10.1042/BSR20182361. Print 2019 Jun 28. Biosci Rep. 2019. PMID: 31142630 Free PMC article.
-
Exploiting Microglial Functions for the Treatment of Glioblastoma.Curr Cancer Drug Targets. 2017;17(3):267-281. doi: 10.2174/1568009616666160813191240. Curr Cancer Drug Targets. 2017. PMID: 27528361 Review.
-
Programmed Cell Death 10 Mediated CXCL2-CXCR2 Signaling in Regulating Tumor-Associated Microglia/Macrophages Recruitment in Glioblastoma.Front Immunol. 2021 May 24;12:637053. doi: 10.3389/fimmu.2021.637053. eCollection 2021. Front Immunol. 2021. PMID: 34108959 Free PMC article.
-
P4HB and PDIA3 are associated with tumor progression and therapeutic outcome of diffuse gliomas.Oncol Rep. 2018 Feb;39(2):501-510. doi: 10.3892/or.2017.6134. Epub 2017 Dec 4. Oncol Rep. 2018. PMID: 29207176 Free PMC article.
-
Insights in the immunobiology of glioblastoma.J Mol Med (Berl). 2020 Jan;98(1):1-10. doi: 10.1007/s00109-019-01835-4. Epub 2019 Oct 24. J Mol Med (Berl). 2020. PMID: 31650201 Review.
Cited by
-
Using AI-Based Evolutionary Algorithms to Elucidate Adult Brain Tumor (Glioma) Etiology Associated with IDH1 for Therapeutic Target Identification.Curr Issues Mol Biol. 2022 Jul 2;44(7):2982-3000. doi: 10.3390/cimb44070206. Curr Issues Mol Biol. 2022. PMID: 35877430 Free PMC article.
-
Protein Disulfide Isomerase A3 (PDIA3): A Pharmacological Target in Glioblastoma?Int J Mol Sci. 2023 Aug 26;24(17):13279. doi: 10.3390/ijms241713279. Int J Mol Sci. 2023. PMID: 37686085 Free PMC article.
-
Pan-Cancer Analysis of PDIA3: Identifying It as a Potential Biomarker for Tumor Prognosis and Immunotherapy.Oxid Med Cell Longev. 2022 Aug 22;2022:9614819. doi: 10.1155/2022/9614819. eCollection 2022. Oxid Med Cell Longev. 2022. PMID: 36046686 Free PMC article.
-
Interactions between microglia and glioma in tumor microenvironment.Front Oncol. 2023 Aug 28;13:1236268. doi: 10.3389/fonc.2023.1236268. eCollection 2023. Front Oncol. 2023. PMID: 37700840 Free PMC article. Review.
-
Protein disulfide-isomerase A4 confers glioblastoma angiogenesis promotion capacity and resistance to anti-angiogenic therapy.J Exp Clin Cancer Res. 2023 Mar 30;42(1):77. doi: 10.1186/s13046-023-02640-1. J Exp Clin Cancer Res. 2023. PMID: 36997943 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

