Targeted Disruption of Mouse Dip2B Leads to Abnormal Lung Development and Prenatal Lethality
- PMID: 33153107
- PMCID: PMC7663123
- DOI: 10.3390/ijms21218223
Targeted Disruption of Mouse Dip2B Leads to Abnormal Lung Development and Prenatal Lethality
Abstract
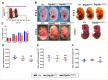
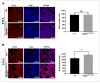
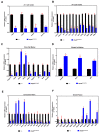
Molecular and anatomical functions of mammalian Dip2 family members (Dip2A, Dip2B and Dip2C) during organogenesis are largely unknown. Here, we explored the indispensable role of Dip2B in mouse lung development. Using a LacZ reporter, we explored Dip2B expression during embryogenesis. This study shows that Dip2B expression is widely distributed in various neuronal, myocardial, endothelial, and epithelial cell types during embryogenesis. Target disruption of Dip2b leads to intrauterine growth restriction, defective lung formation and perinatal mortality. Dip2B is crucial for late lung maturation rather than early-branching morphogenesis. The morphological analysis shows that Dip2b loss leads to disrupted air sac formation, interstitium septation and increased cellularity. In BrdU incorporation assay, it is shown that Dip2b loss results in increased cell proliferation at the saccular stage of lung development. RNA-seq analysis reveals that 1431 genes are affected in Dip2b deficient lungs at E18.5 gestation age. Gene ontology analysis indicates cell cycle-related genes are upregulated and immune system related genes are downregulated. KEGG analysis identifies oxidative phosphorylation as the most overrepresented pathways along with the G2/M phase transition pathway. Loss of Dip2b de-represses the expression of alveolar type I and type II molecular markers. Altogether, the study demonstrates an important role of Dip2B in lung maturation and survival.
Keywords: Dip2b; LacZ expression; RNA sequencing; fetal lung development; growth restriction; mice; prenatal lethality.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures




References
-
- Tanaka M., Murakami K., Ozaki S., Imura Y., Tong X.-P., Watanabe T., Sawaki T., Kawanami T., Kawabata D., Fujii T., et al. DIP2 disco-interacting protein 2 homolog A (Drosophila) is a candidate receptor for follistatin-related protein/follistatin-like 1—Analysis of their binding with TGF-β superfamily proteins. FEBS J. 2010;277:4278–4289. doi: 10.1111/j.1742-4658.2010.07816.x. - DOI - PubMed
-
- Mukhopadhyay M., Pelka P., DeSousa D., Kablar B., Schindler A., Rudnicki M.A., Campos A.R. Cloning, genomic organization and expression pattern of a novel Drosophila gene, the disco-interacting protein 2 (dip2), and its murine homolog. Gene. 2002;293:59–65. doi: 10.1016/S0378-1119(02)00694-7. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

