Characterization of Mutational Status, Spheroid Formation, and Drug Response of a New Genomically-Stable Human Ovarian Clear Cell Carcinoma Cell Line, 105C
- PMID: 33153119
- PMCID: PMC7693681
- DOI: 10.3390/cells9112408
Characterization of Mutational Status, Spheroid Formation, and Drug Response of a New Genomically-Stable Human Ovarian Clear Cell Carcinoma Cell Line, 105C
Abstract
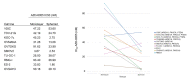
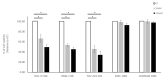
Ovarian clear cell carcinoma (OCCC) is a rare subtype of gynecological cancer for which well-characterized and authenticated model systems are scarce. We provide an extensive characterization of '105C', a cell line generated from an adenocarcinoma of the clear cell histotype using targeted next-generation sequencing, cytogenetic microarrays, along with analyses of AKT/mTOR signaling. We report that that the 105C cell line is a bona fide OCCC cell line, carrying PIK3CA, PTEN, and ARID1A gene mutations, consistent with OCCC, yet maintain a stable genome as reflected by low copy number variation. Unlike KOC-7c, TOV-21G, and RMG-V OCCC lines also mutated for the above genes, the 105C cells do not carry mutations in mismatch repair genes. Importantly, we show that 105C cells exhibit greater resistance to mTOR inhibition and carboplatin treatment compared to 9 other OCCC cell lines in 3D spheroid cultures. This resistance may be attributed to 105C cells remaining dormant in suspension culture which surprisingly, contrasts with several other OCCC lines which continue to proliferate in long-term suspension culture. 105C cells survive xenotransplantation but do not proliferate and metastasize. Collectively, we show that the 105C OCCC cell line exhibits unique properties useful for the pre-clinical investigation of OCCC pathobiology.
Keywords: OCCC; cancer; cell line; clear cell; epithelial ovarian cancer; mTOR inhibitor; ovarian; spheroid.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Transcriptomic Analyses of Ovarian Clear Cell Carcinoma Spheroids Reveal Distinct Proliferative Phenotypes and Therapeutic Vulnerabilities.Cells. 2025 May 27;14(11):785. doi: 10.3390/cells14110785. Cells. 2025. PMID: 40497961 Free PMC article.
-
CCNE1 Is a Putative Therapeutic Target for ARID1A-Mutated Ovarian Clear Cell Carcinoma.Int J Mol Sci. 2021 May 30;22(11):5869. doi: 10.3390/ijms22115869. Int J Mol Sci. 2021. PMID: 34070839 Free PMC article.
-
Genome-wide DNA methylation in relation to ARID1A deficiency in ovarian clear cell carcinoma.J Transl Med. 2024 Jun 10;22(1):556. doi: 10.1186/s12967-024-05311-7. J Transl Med. 2024. PMID: 38858765 Free PMC article.
-
Clinical analysis and literature review of a case of ovarian clear cell carcinoma with PIK3CA gene mutation: A case report.Medicine (Baltimore). 2022 Sep 16;101(37):e30666. doi: 10.1097/MD.0000000000030666. Medicine (Baltimore). 2022. PMID: 36123851 Free PMC article. Review.
-
Precision medicine for ovarian clear cell carcinoma based on gene alterations.Int J Clin Oncol. 2020 Mar;25(3):419-424. doi: 10.1007/s10147-020-01622-z. Epub 2020 Feb 4. Int J Clin Oncol. 2020. PMID: 32020380 Review.
Cited by
-
Transcriptomic Analyses of Ovarian Clear Cell Carcinoma Spheroids Reveal Distinct Proliferative Phenotypes and Therapeutic Vulnerabilities.Cells. 2025 May 27;14(11):785. doi: 10.3390/cells14110785. Cells. 2025. PMID: 40497961 Free PMC article.
-
miRNA-Dependent Regulation of AKT1 Phosphorylation.Cells. 2022 Feb 26;11(5):821. doi: 10.3390/cells11050821. Cells. 2022. PMID: 35269443 Free PMC article.
-
Comparative analysis of response to treatments and molecular features of tumor-derived organoids versus cell lines and PDX derived from the same ovarian clear cell carcinoma.J Exp Clin Cancer Res. 2023 Oct 7;42(1):260. doi: 10.1186/s13046-023-02809-8. J Exp Clin Cancer Res. 2023. PMID: 37803448 Free PMC article.
-
Assessing the antitumor effects of metformin on ovarian clear cell carcinoma.Hum Cell. 2024 Sep;37(5):1462-1474. doi: 10.1007/s13577-024-01116-4. Epub 2024 Aug 8. Hum Cell. 2024. PMID: 39115639
-
CRISPR/Cas9 Screening for Identification of Genes Required for the Growth of Ovarian Clear Cell Carcinoma Cells.Curr Issues Mol Biol. 2022 Apr 7;44(4):1587-1596. doi: 10.3390/cimb44040108. Curr Issues Mol Biol. 2022. PMID: 35723366 Free PMC article.
References
-
- Hermens M., van Altena A.M., Nieboer T.E., Schoot B.C., van Vliet H.A.A.M., Siebers A.G., Bekkers R.L.M. Incidence of endometrioid and clear-cell ovarian cancer in histological proven endometriosis: The ENOCA population-based cohort study. Am. J. Obstet. Gynecol. 2020;223:107.e1–107.e11. doi: 10.1016/j.ajog.2020.01.041. - DOI - PubMed
-
- Köbel M., Kalloger S.E., Huntsman D.G., Santos J.L., Swenerton K.D., Seidman J.D., Gilks C.B., Cheryl B. Ovarian Cancer Outcomes Unit of the British Columbia Cancer Agency VB. Differences in tumor type in low-stage versus high-stage ovarian carcinomas. Int. J. Gynecol. Pathol. 2010;29:203–211. doi: 10.1097/PGP.0b013e3181c042b6. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

