Coronaviruses: Innate Immunity, Inflammasome Activation, Inflammatory Cell Death, and Cytokines
- PMID: 33153908
- PMCID: PMC7561287
- DOI: 10.1016/j.it.2020.10.005
Coronaviruses: Innate Immunity, Inflammasome Activation, Inflammatory Cell Death, and Cytokines
Abstract
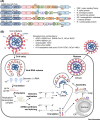
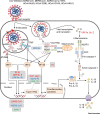
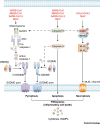
The innate immune system acts as the first line of defense against pathogens, including coronaviruses (CoVs). Severe acute respiratory syndrome (SARS)-CoV and Middle East respiratory syndrome (MERS)-CoV are epidemic zoonotic CoVs that emerged at the beginning of the 21st century. The recently emerged virus SARS-CoV-2 is a novel strain of CoV that has caused the coronavirus 2019 (COVID-19) pandemic. Scientific advancements made by studying the SARS-CoV and MERS-CoV outbreaks have provided a foundation for understanding pathogenesis and innate immunity against SARS-CoV-2. In this review, we focus on our present understanding of innate immune responses, inflammasome activation, inflammatory cell death pathways, and cytokine secretion during SARS-CoV, MERS-CoV, and SARS-CoV-2 infection. We also discuss how the pathogenesis of these viruses influences these biological processes.
Keywords: COVID-19; MERS-CoV; MHV; PANoptosis; PANoptosome; RNA virus; SARS-CoV; SARS-CoV-2; cell death; coronavirus; cytokines; inflammasome; inflammation; innate immunity.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

