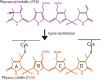
MpeV is a lyase isomerase that ligates a doubly linked phycourobilin on the β-subunit of phycoerythrin I and II in marine Synechococcus
- PMID: 33154169
- PMCID: PMC7948978
- DOI: 10.1074/jbc.RA120.015289
MpeV is a lyase isomerase that ligates a doubly linked phycourobilin on the β-subunit of phycoerythrin I and II in marine Synechococcus
Abstract
Synechococcus cyanobacteria are widespread in the marine environment, as the extensive pigment diversity within their light-harvesting phycobilisomes enables them to utilize various wavelengths of light for photosynthesis. The phycobilisomes of Synechococcus sp. RS9916 contain two forms of the protein phycoerythrin (PEI and PEII), each binding two chromophores, green-light absorbing phycoerythrobilin and blue-light absorbing phycourobilin. These chromophores are ligated to specific cysteines via bilin lyases, and some of these enzymes, called lyase isomerases, attach phycoerythrobilin and simultaneously isomerize it to phycourobilin. MpeV is a putative lyase isomerase whose role in PEI and PEII biosynthesis is not clear. We examined MpeV in RS9916 using recombinant protein expression, absorbance spectroscopy, and tandem mass spectrometry. Our results show that MpeV is the lyase isomerase that covalently attaches a doubly linked phycourobilin to two cysteine residues (C50, C61) on the β-subunit of both PEI (CpeB) and PEII (MpeB). MpeV activity requires that CpeB or MpeB is first chromophorylated by the lyase CpeS (which adds phycoerythrobilin to C82). Its activity is further enhanced by CpeZ (a homolog of a chaperone-like protein first characterized in Fremyella diplosiphon). MpeV showed no detectable activity on the α-subunits of PEI or PEII. The mechanism by which MpeV links the A and D rings of phycourobilin to C50 and C61 of CpeB was also explored using site-directed mutants, revealing that linkage at the A ring to C50 is a critical step in chromophore attachment, isomerization, and stability. These data provide novel insights into β-PE biosynthesis and advance our understanding of the mechanisms guiding lyase isomerases.
Keywords: bilin lyase; cyanobacteria; lyase isomerase; phycobilisome; phycoerythrobilin; phycourobilin; posttranslational modification.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures




Similar articles
-
The phycoerythrobilin isomerization activity of MpeV in Synechococcus sp. WH8020 is prevented by the presence of a histidine at position 141 within its phycoerythrin-I β-subunit substrate.Front Microbiol. 2022 Nov 15;13:1011189. doi: 10.3389/fmicb.2022.1011189. eCollection 2022. Front Microbiol. 2022. PMID: 36458192 Free PMC article.
-
CpeY is a phycoerythrobilin lyase for cysteine 82 of the phycoerythrin I α-subunit in marine Synechococcus.Biochim Biophys Acta Bioenerg. 2020 Aug 1;1861(8):148215. doi: 10.1016/j.bbabio.2020.148215. Epub 2020 Apr 29. Biochim Biophys Acta Bioenerg. 2020. PMID: 32360311
-
CpeF is the bilin lyase that ligates the doubly linked phycoerythrobilin on β-phycoerythrin in the cyanobacterium Fremyella diplosiphon.J Biol Chem. 2019 Mar 15;294(11):3987-3999. doi: 10.1074/jbc.RA118.007221. Epub 2019 Jan 22. J Biol Chem. 2019. PMID: 30670589 Free PMC article.
-
Phycobiliproteins in Prochlorococcus marinus: biosynthesis of pigments and their assembly into proteins.Eur J Cell Biol. 2010 Dec;89(12):1005-10. doi: 10.1016/j.ejcb.2010.06.017. Epub 2010 Aug 17. Eur J Cell Biol. 2010. PMID: 20724022 Review.
-
Prospects of phycoerythrin: Structural features, antioxidation and applications in food.Food Chem. 2025 Jan 15;463(Pt 4):141425. doi: 10.1016/j.foodchem.2024.141425. Epub 2024 Sep 25. Food Chem. 2025. PMID: 39348767 Review.
Cited by
-
Temporal and Spatial Dynamics of Synechococcus Clade II and Other Microbes in the Eutrophic Subtropical San Diego Bay.Environ Microbiol. 2025 Feb;27(2):e70043. doi: 10.1111/1462-2920.70043. Environ Microbiol. 2025. PMID: 39900485 Free PMC article.
-
Red algae acclimate to low light by modifying phycobilisome composition to maintain efficient light harvesting.BMC Biol. 2022 Dec 27;20(1):291. doi: 10.1186/s12915-022-01480-3. BMC Biol. 2022. PMID: 36575464 Free PMC article.
-
Phycobiliproteins-A Family of Algae-Derived Biliproteins: Productions, Characterization and Pharmaceutical Potentials.Mar Drugs. 2022 Jul 9;20(7):450. doi: 10.3390/md20070450. Mar Drugs. 2022. PMID: 35877743 Free PMC article. Review.
-
Diversity and Evolution of Pigment Types in Marine Synechococcus Cyanobacteria.Genome Biol Evol. 2022 Apr 10;14(4):evac035. doi: 10.1093/gbe/evac035. Genome Biol Evol. 2022. PMID: 35276007 Free PMC article.
-
Phycobiliproteins: Structural aspects, functional characteristics, and biotechnological perspectives.Comput Struct Biotechnol J. 2022 Feb 23;20:1506-1527. doi: 10.1016/j.csbj.2022.02.016. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35422968 Free PMC article. Review.
References
-
- Flombaum P., Gallegos J.L., Gordillo R.A., Rincon J., Zabala L.L., Jiao N., Karl D.M., Li W.K., Lomas M.W., Veneziano D., Vera C.S., Vrugt J.A., Martiny A.C. Present and future global distributions of the marine cyanobacteria Prochlorococcus and Synechococcus. Proc. Natl. Acad. Sci. U. S. A. 2013;110:9824–9829. - PMC - PubMed
-
- Glazer A.N. Adaptive variation in phycobilisome structure. Adv. Mol. Cell Biol. 1994;10:119–149.
-
- Sanfilippo J.E., Garczarek L., Partensky F., Kehoe D.M. Chromatic acclimation in cyanobacteria: a diverse and widespread process for optimizing photosynthesis. Ann. Rev. Microbiol. 2019;73:407–433. - PubMed
-
- Schluchter W.M., Shen G., Alvey R.M., Biswas A., Saunee N.A., Williams S.R., Miller C.A., Bryant D.A. Phycobiliprotein biosynthesis in cyanobacteria: structure and function of enzymes involved in post-translational modification. Adv. Exp. Med. Biol. 2010;675:211–228. - PubMed
-
- Glazer A.N. Phycobiliproteins - a family of valuable, widely used fluorophores. J. Appl. Phycol. 1994;6:105–112.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

