Deep Learning-Based Segmentation and Quantification in Experimental Kidney Histopathology
- PMID: 33154175
- PMCID: PMC7894663
- DOI: 10.1681/ASN.2020050597
Deep Learning-Based Segmentation and Quantification in Experimental Kidney Histopathology
Abstract
Background: Nephropathologic analyses provide important outcomes-related data in experiments with the animal models that are essential for understanding kidney disease pathophysiology. Precision medicine increases the demand for quantitative, unbiased, reproducible, and efficient histopathologic analyses, which will require novel high-throughput tools. A deep learning technique, the convolutional neural network, is increasingly applied in pathology because of its high performance in tasks like histology segmentation.
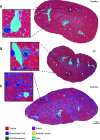
Methods: We investigated use of a convolutional neural network architecture for accurate segmentation of periodic acid-Schiff-stained kidney tissue from healthy mice and five murine disease models and from other species used in preclinical research. We trained the convolutional neural network to segment six major renal structures: glomerular tuft, glomerulus including Bowman's capsule, tubules, arteries, arterial lumina, and veins. To achieve high accuracy, we performed a large number of expert-based annotations, 72,722 in total.
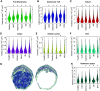
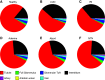
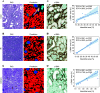
Results: Multiclass segmentation performance was very high in all disease models. The convolutional neural network allowed high-throughput and large-scale, quantitative and comparative analyses of various models. In disease models, computational feature extraction revealed interstitial expansion, tubular dilation and atrophy, and glomerular size variability. Validation showed a high correlation of findings with current standard morphometric analysis. The convolutional neural network also showed high performance in other species used in research-including rats, pigs, bears, and marmosets-as well as in humans, providing a translational bridge between preclinical and clinical studies.
Conclusions: We developed a deep learning algorithm for accurate multiclass segmentation of digital whole-slide images of periodic acid-Schiff-stained kidneys from various species and renal disease models. This enables reproducible quantitative histopathologic analyses in preclinical models that also might be applicable to clinical studies.
Keywords: animal model; digital pathology; histopathology; segmentation.
Copyright © 2021 by the American Society of Nephrology.
Figures





References
-
- Robboy SJ, Weintraub S, Horvath AE, Jensen BW, Alexander CB, Fody EP, et al. .: Pathologist workforce in the United States: I. Development of a predictive model to examine factors influencing supply. Arch Pathol Lab Med 137: 1723–1732, 2013 - PubMed
-
- LeCun Y, Bengio Y, Hinton G: Deep learning. Nature 521: 436–444, 2015 - PubMed
-
- Boor P: Artificial intelligence in nephropathology. Nat Rev Nephrol 16: 4–6, 2020 - PubMed
-
- Sirinukunwattana K, Ahmed Raza SE, Yee-Wah Tsang, Snead DRJ, Cree IA, Rajpoot NM: Locality sensitive deep learning for detection and classification of nuclei in routine colon cancer histology images. IEEE Trans Med Imaging 35: 1196–1206, 2016 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

