Spatially and cell-type resolved quantitative proteomic atlas of healthy human skin
- PMID: 33154365
- PMCID: PMC7645789
- DOI: 10.1038/s41467-020-19383-8
Spatially and cell-type resolved quantitative proteomic atlas of healthy human skin
Abstract
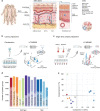
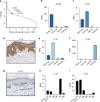
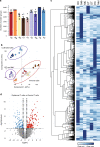
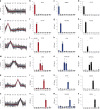
Human skin provides both physical integrity and immunological protection from the external environment using functionally distinct layers, cell types and extracellular matrix. Despite its central role in human health and disease, the constituent proteins of skin have not been systematically characterized. Here, we combine advanced tissue dissection methods, flow cytometry and state-of-the-art proteomics to describe a spatially-resolved quantitative proteomic atlas of human skin. We quantify 10,701 proteins as a function of their spatial location and cellular origin. The resulting protein atlas and our initial data analyses demonstrate the value of proteomics for understanding cell-type diversity within the skin. We describe the quantitative distribution of structural proteins, known and previously undescribed proteins specific to cellular subsets and those with specialized immunological functions such as cytokines and chemokines. We anticipate that this proteomic atlas of human skin will become an essential community resource for basic and translational research ( https://skin.science/ ).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Heath WR, Carbone FR. The skin-resident and migratory immune system in steady state and memory: innate lymphocytes, dendritic cells and T cells. Nat. Immunol. 2013;14:978–985. - PubMed
-
- Bos JD, et al. The skin immune system (SIS): distribution and immunophenotype of lymphocyte subpopulations in normal human skin. J. Invest. Dermatol. 1987;88:569–573. - PubMed
-
- Clark RA, et al. The vast majority of CLA+ T cells are resident in normal skin. J. Immunol. 2006;176:4431–4439. - PubMed
-
- Dengjel J, Bruckner-Tuderman L, Nyström A. Skin proteomics—analysis of the extracellular matrix in health and disease. Expert Rev. Proteomics. 2020;17:377–391. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

