Potential involvement of Streptococcus mutans possessing collagen binding protein Cnm in infective endocarditis
- PMID: 33154489
- PMCID: PMC7645802
- DOI: 10.1038/s41598-020-75933-6
Potential involvement of Streptococcus mutans possessing collagen binding protein Cnm in infective endocarditis
Abstract
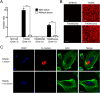
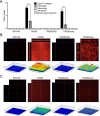
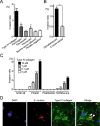
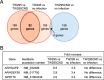
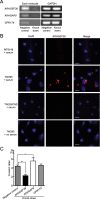
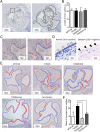
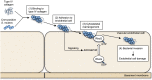
Streptococcus mutans, a significant contributor to dental caries, is occasionally isolated from the blood of patients with infective endocarditis. We previously showed that S. mutans strains expressing collagen-binding protein (Cnm) are present in the oral cavity of approximately 10-20% of humans and that they can effectively invade human umbilical vein endothelial cells (HUVECs). Here, we investigated the potential molecular mechanisms of HUVEC invasion by Cnm-positive S. mutans. The ability of Cnm-positive S. mutans to invade HUVECs was significantly increased by the presence of serum, purified type IV collagen, and fibrinogen (p < 0.001). Microarray analyses of HUVECs infected by Cnm-positive or -negative S. mutans strains identified several transcripts that were differentially upregulated during invasion, including those encoding the small G protein regulatory proteins ARHGEF38 and ARHGAP9. Upregulation of these proteins occurred during invasion only in the presence of serum. Knockdown of ARHGEF38 strongly reduced HUVEC invasion by Cnm-positive S. mutans. In a rat model of infective endocarditis, cardiac endothelial cell damage was more prominent following infection with a Cnm-positive strain compared with a Cnm-negative strain. These results suggest that the type IV collagen-Cnm-ARHGEF38 pathway may play a crucial role in the pathogenesis of infective endocarditis.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Roberts GJ, Lucas VS, Omar J. Bacterial endocarditis and orthodontics. J. R. Coll. Surg. Edinb. 2000;45:141–145. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

