Identification of early liver toxicity gene biomarkers using comparative supervised machine learning
- PMID: 33154507
- PMCID: PMC7645727
- DOI: 10.1038/s41598-020-76129-8
Identification of early liver toxicity gene biomarkers using comparative supervised machine learning
Abstract
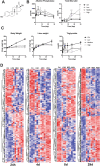
Screening agrochemicals and pharmaceuticals for potential liver toxicity is required for regulatory approval and is an expensive and time-consuming process. The identification and utilization of early exposure gene signatures and robust predictive models in regulatory toxicity testing has the potential to reduce time and costs substantially. In this study, comparative supervised machine learning approaches were applied to the rat liver TG-GATEs dataset to develop feature selection and predictive testing. We identified ten gene biomarkers using three different feature selection methods that predicted liver necrosis with high specificity and selectivity in an independent validation dataset from the Microarray Quality Control (MAQC)-II study. Nine of the ten genes that were selected with the supervised methods are involved in metabolism and detoxification (Car3, Crat, Cyp39a1, Dcd, Lbp, Scly, Slc23a1, and Tkfc) and transcriptional regulation (Ablim3). Several of these genes are also implicated in liver carcinogenesis, including Crat, Car3 and Slc23a1. Our biomarker gene signature provides high statistical accuracy and a manageable number of genes to study as indicators to potentially accelerate toxicity testing based on their ability to induce liver necrosis and, eventually, liver cancer.
Conflict of interest statement
There are competing interests between the authors (ZME, RB) and Corteva Agrisciences (NE, KJ); specifically the research was supported by Corteva Agrisciences. Other authors do not declare competing interests.
Figures


Similar articles
-
Blood-based gene-expression biomarkers identification for the non-invasive diagnosis of Parkinson's disease using two-layer hybrid feature selection.Gene. 2022 May 20;823:146366. doi: 10.1016/j.gene.2022.146366. Epub 2022 Feb 22. Gene. 2022. PMID: 35202733
-
Unraveling the mechanisms underlying drug-induced cholestatic liver injury: identifying key genes using machine learning techniques on human in vitro data sets.Arch Toxicol. 2023 Nov;97(11):2969-2981. doi: 10.1007/s00204-023-03583-4. Epub 2023 Aug 21. Arch Toxicol. 2023. PMID: 37603094 Free PMC article.
-
A novel transcriptomics based in vitro method to compare and predict hepatotoxicity based on mode of action.Toxicology. 2015 Feb 3;328:29-39. doi: 10.1016/j.tox.2014.11.008. Epub 2014 Dec 2. Toxicology. 2015. PMID: 25475144
-
The discovery and development of proteomic safety biomarkers for the detection of drug-induced liver toxicity.Toxicol Appl Pharmacol. 2010 May 15;245(1):134-42. doi: 10.1016/j.taap.2010.02.011. Epub 2010 Feb 26. Toxicol Appl Pharmacol. 2010. PMID: 20219512 Review.
-
Iterative processes: a review of semi-supervised machine learning in rehabilitation science.Disabil Rehabil Assist Technol. 2020 Jul;15(5):515-520. doi: 10.1080/17483107.2019.1604831. Epub 2019 Jul 8. Disabil Rehabil Assist Technol. 2020. PMID: 31282778 Review.
Cited by
-
A novel support vector machine-based 1-day, single-dose prediction model of genotoxic hepatocarcinogenicity in rats.Arch Toxicol. 2024 Aug;98(8):2711-2730. doi: 10.1007/s00204-024-03755-w. Epub 2024 May 18. Arch Toxicol. 2024. PMID: 38762666
-
PFAS and their association with the increased risk of cardiovascular disease in postmenopausal women.Toxicol Sci. 2024 Aug 1;200(2):312-323. doi: 10.1093/toxsci/kfae065. Toxicol Sci. 2024. PMID: 38758093 Free PMC article.
-
Artificial Intelligence in Liver Diseases: Recent Advances.Adv Ther. 2024 Mar;41(3):967-990. doi: 10.1007/s12325-024-02781-5. Epub 2024 Jan 29. Adv Ther. 2024. PMID: 38286960 Review.
-
Progress in toxicogenomics to protect human health.Nat Rev Genet. 2025 Feb;26(2):105-122. doi: 10.1038/s41576-024-00767-1. Epub 2024 Sep 2. Nat Rev Genet. 2025. PMID: 39223311 Review.
-
AI-driven Discovery of Morphomolecular Signatures in Toxicology.bioRxiv [Preprint]. 2024 Jul 23:2024.07.19.604355. doi: 10.1101/2024.07.19.604355. bioRxiv. 2024. PMID: 39091765 Free PMC article. Preprint.
References
-
- Laura Suter-Dick FP. Predictive Toxicology. New York: Springer; 2014.
-
- Längkvist M, Karlsson L, Loutfi A. A review of unsupervised feature learning and deep learning for time-series modeling. Pattern Recogn. Lett. 2014;42:11–24. doi: 10.1016/j.patrec.2014.01.008. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

